Abstract
Conventional influenza vaccines are based on predicting the circulating viruses year by year, conferring limited effectiveness since the antigenicity of vaccine strains does not always match the circulating viruses. This necessitates development of universal influenza vaccines that provide broader and lasting protection against pan-influenza viruses. The discovery of the highly conserved immunogens (epitopes) of influenza viruses provides attractive targets for universal vaccine design. Here we review the current understanding with broadly protective immunogens (epitopes) and discuss several important considerations to achieve the goal of universal influenza vaccines.
Similar content being viewed by others
Introduction
More than 100 years past the catastrophic 1918 Spanish influenza pandemic, influenza viruses remain a constant global health threat. Three types of influenza viruses infect humans: type A (influenza A virus, IAV), type B (IBV) and type C (ICV). Two IAV subtypes (H3N2 and H1N1pdm09) and two IBV lineages (Yamagata and Victoria) co-circulate annually, causing seasonal epidemics and up to 650,000 respiratory deaths globally, while ICV is detected less frequently and usually causes mild infections (Krammer et al. 2018; WHO 2018). In addition, antigenically novel IAVs generated by reassortment of the segmented genome can occasionally infect humans with high rates of morbidity and mortality, as well as potential pandemic risk (Krammer et al. 2018; WHO 2018).
Vaccines provide cost-effective protection against influenza. Currently, seasonal influenza virus vaccines predominantly induce antibody responses against the hemagglutinin (HA), one of the two major surface glycoproteins of the virus (Krammer 2019). However, the majority of vaccine-induced antibodies are directed against the highly plastic head region of HA and are strain-specific (Caton et al. 1982; Heaton et al. 2013). Amino acid residues on the surface of this immunodominant head region vary substantially among different strains and change continuously (referred to as antigenic drift), leading to new circulating virus strains (Wang and Palese 2011). Therefore, current influenza vaccines have to be reformulated each year based on surveillance and prediction, which is cumbersome, and their effectiveness is highly variable depending on the accurate forecasts of the circulating strains in each year (de Jong et al. 2000; Gerdil 2003). Moreover, the protection conferred by seasonal influenza vaccines does not cover emerging pandemic influenza virus strains.
These limitations of current vaccines emphasize the need to develop novel “universal” or “broadly protective” influenza vaccines. An ideal universal vaccine should cover all influenza A and influenza B viruses independent of antigenic drift or HA/neuraminidase (NA) subtype. In contrast, a broadly protective vaccine would cover a large subset of influenza viruses, for example, all human seasonal influenza virus strains. These two types of vaccines are generally referred to as “universal vaccines” (Memoli et al. 2019).
Most universal vaccines in development aim at inducing broadly protective antibody responses, while some others focus on inducing T cell responses alternatively (Nachbagauer and Krammer 2017). In either case, the conserved immunogens of the virus need to be targeted, and the mechanism of protection mainly depends on the immunogens of choice (Nachbagauer and Krammer 2017; Asthagiri Arunkumar et al. 2019). This review will discuss the conserved antigenic regions/epitopes of influenza viruses that can facilitate immunogen design and contribute to the development of universal influenza vaccines.
Surface Proteins and Antibody Responses
Induction of protective antibody responses against influenza viruses requires host recognition of the viral surface proteins, including HA, NA and matrix protein 2 (M2) (Fig. 1A). Thus the discovery of new broadly protective antibodies and related conserved epitopes can greatly accelerate design and implementation of universal vaccines (Deng and Wang 2018).
Hemagglutinin
HA is assembled as homotrimers consisting of two domains, the membrane-distal globular head domain and the membrane-proximal helix-rich stalk domain (Fig. 1B). Based on the antigenicity, HA can be divided into 18 subtypes, which fall into 2 groups, including group 1 (H1, H2, H5, H6, H8, H9, H11, H12, H13, H16, H17, H18) and group 2 (H3, H4, H7, H10, H14, H15) (Du et al. 2019). As mentioned above, HA head domain is the major target of the antibody response following vaccination with seasonal influenza vaccines, however its high plasticity makes it easy for the virus to escape immune pressure (Heaton et al. 2013). Despite of this, monoclonal antibodies (mAbs) that cross-react between the head domains of different HA subtypes have been reported (Ekiert et al. 2012; Krause et al. 2012; Lee et al. 2012; Boonsathorn et al. 2014; Lee et al. 2014). These cross-reactive head antibodies seem rare, but they are interesting since they suggest the existence of cross-reactive epitopes in the head domains.
In contrast, HA stalk is the least variable region of HA, and great efforts have been focused on the development of universal influenza vaccines that depend on protective epitopes in this domain (Krammer and Palese 2015; Wu and Wilson 2018). Monoclonal antibodies against this domain can broadly neutralize different subtypes of group 1 (Okuno et al. 1993; Ekiert et al. 2009; Sui et al. 2009) or group 2 viruses (Ekiert et al. 2011) or even both (Corti et al. 2011). Interestingly, although some stalk-antibodies show lower or even none neutralizing potency in vitro, they can render robust protection against challenges with divergent influenza viruses in vivo (Dreyfus et al. 2012; DiLillo et al. 2014; He et al. 2015), most likely by Fc-mediated mechanisms like antibody dependent cell-mediated cytotoxicity (ADCC) and complement-dependent cytotoxicity (CDC) (Terajima et al. 2011; Jegaskanda et al. 2013a, b; Miller et al. 2013; DiLillo et al. 2014; Florek et al. 2014; Jegaskanda et al. 2014).
Glycosylation and conformational mobility (shape-shifting) of surface glycoproteins can help viruses to mask sensitive epitopes and evade the immune system. Interestingly, two latest studies revealed that glycan repositioning within either head or stem domains of HA can facilitate the elicitation of protective cross-reactive antibody responses. Huang et al. reported that removing glycosylation at residue 144 and deletion of lysine at position 147 can synergize to elicit broadly-reactive H1N1 influenza antibodies (Huang et al. 2019). A study by Boyoglu-Barnum et al. showed that introduction of the group 2 glycan at Asn38HA1 to a group 1 stem-nanoparticle can broaden antibody responses to cross-react with group 2 HAs (Boyoglu-Barnum et al. 2020). Although the precise sites that are vulnerable by glycan repositioning are not identified yet, they may provide novel target epitopes to achieve universal protection.
More recently, a novel conserved epitope at the HA head domain trimer interface was reported (Bajic et al. 2019; Bangaru et al. 2019; Watanabe et al. 2019). Primarily, the epitope is occluded at the contact surface between HA head domains, while the reversible HA “breathing” conformational dynamics can cause exposure of the interface. Antibodies targeting this cryptic epitope were shown to be protective against broad-spectrum subtypes of IAVs, suggesting the HA head interface is a new potential immunogen component for influenza universal vaccine design (Bajic et al. 2019; Bangaru et al. 2019; Watanabe et al. 2019; Wu and Gao 2019).
Neuraminidase
NA is the second most abundant glycoprotein on the surface of influenza A and B viruses, and plays an essential role during virus replication by facilitating release of progeny virions (Fig. 1B) (Du et al. 2019). It is well known that NA antibodies can protect against infection of influenza viruses (Schulman et al. 1968; Murphy et al. 1972; Monto and Kendal 1973; Doyle et al. 2013; Wan et al. 2013; Wilson et al. 2016; Wohlbold et al. 2017), by preventing virus budding and egress from infected cells, as well as ADCC and CDC (Wan et al. 2013; Krammer and Palese 2015; Wohlbold et al. 2017). NA consists of 11 subtypes (N1 to N11), and a series of “broadly” cross-reactive mAbs of NA have been isolated (Wan et al. 2013; Wilson et al. 2016; Wohlbold et al. 2017; Chen et al. 2018). Of note, most of these cross-reactive NA mAbs are limited to recognize heterologous virus strains of the same NA subtype, suggesting existence of conserved subtype-specific NA epitopes (Wan et al. 2013; Wilson et al. 2016; Wohlbold et al. 2017; Chen et al. 2018).
Interestingly, Doyle et al. reported a unique universally conserved sequence (amino acids 222–230, located in the vicinity of enzymatic active site) amongst all IAV NA subtypes, and a mAb targeting this specific epitope was demonstrated to inhibit all NA subtypes and protect mice from lethal doses of mouse-adapted H1N1 and H3N2 (Doyle et al. 2013). More recently, Stadlbauer et al. identified three broadly-protective mAbs targeting the NA active site that is highly conserved among all NA subtypes (Stadlbauer et al. 2019). These conserved NA epitopes, either subtype specific or universal, may be attractive immunological target for universal influenza vaccine design.
Matrix Protein 2
M2 of IAV (AM2) is a type III integral membrane protein, consisting of an N-terminal extracellular region (M2e), transmembrane region, and C-terminal cytoplasmic tail region (Fig. 1B) (Tobler et al. 1999). The M2e is a 24-residue peptide and is highly conserved among different IAV subtypes (Deng et al. 2015). Although the natural M2e has low immunogenicity and abundance, a M2e-specific and protective monoclonal antibody 14C2 has been isolated early (Zebedee and Lamb 1989; Treanor et al. 1990). Therefore this region has since been extensively evaluated as a promising immunogen for universal influenza vaccines (Farahmand et al. 2019; Yao et al. 2019).
BM2 of IBV is a functional homolog of AM2 but shares little sequence identity (Wanitchang et al. 2016; Mandala et al. 2019), and most mAbs targeting M2e are IAV-wide. Despite a mAb AS2 that recognizes the conservative N-terminus of M2 (amino acids 2–10) has been identified to possess neutralizing activity against both IAV and IBV in vitro (Liu et al. 2003), no evidence showed that immunity with current universal influenza vaccine candidates based on M2e of IAV can suppress IBV replication (Mezhenskaya et al. 2019).
Internal Proteins and T-cell Responses
CD8+ T cells can detect and kill virus-infected cells by recognizing viral protein-derived peptides (epitopes) presented by major histocompatibility complex class I (MHC-I) on the cell surface (Fig. 2A). While CD8+ T cell-based immunity does not prevent infection, it can facilitate viral clearance and reduce the severity of disease (McMichael et al. 1983; Sridhar et al. 2013).
T-cell response and influenza viral internal proteins. A CD8+ T cell response. Virus-derived peptide antigens presented by major histocompatibility complex-I (MHC-1) at surface of virus infected cells can be recognized by CD8+ T cells through specific T cell receptors (TCRs), resulting in lysis of target cells. B A model of influenza viral particle with internal proteins. M1, Matrix protein 1; NP, nucleoprotein; PB1, polymerase basic-1. *, CD8+ T cell targets from IBV HA and NS (nonstructural protein 1) proteins also have been reported.
The current inactivated influenza vaccine formulation does not boost T cell response (Koutsakos et al. 2018). Nevertheless cytotoxic CD8+ T cells specific to the conserved viral epitopes have been evidenced to provide cross-protection across IAV, IBV and ICV (McMichael et al. 1983; Greenbaum et al. 2009; Gras et al. 2010; Sridhar et al. 2013; Quinones-Parra et al. 2014; Hayward et al. 2015; Wang et al. 2015, 2018; Koutsakos et al. 2019). Moreover, pre-existing T cell responses have been demonstrated to correlate with protection from influenza disease in humans by epidemiological studies (Epstein 2006; Hayward et al. 2015). The antigenic origin of these broadly cross-reactive epitopes is therefore of great interest in the development of CD8+ T cell-based universal influenza vaccines. It is believed that CD8+ T cell cross-reactivity is provided by the peptides derived from internal influenza proteins, mainly but not limited to nucleoprotein (NP) and matrix protein 1 (M1) (Fig. 2B) (Koutsakos et al. 2019).
In contrast to HA and NA, the internal influenza virus proteins are relatively well conserved and display limited antigenic diversity (Bui et al. 2007). A number of vaccine candidates have been developed to induce T cell responses, by intracellularly expressing NP and M1 using replication deficient viral vectors like Chimpanzee Adenovirus Oxford 1 (ChAdOx1) or Modified Vaccinia Ankara (MVA) (Berthoud et al. 2011; Lillie et al. 2012; Lambe et al. 2013; Antrobus et al. 2014a, b).
Interestingly, a recent study identified a universal PB1 epitope (PB1413), which is derived from a core motif (residues 406–422 of IAV-PB1 protein) present in the viral RNA-dependent polymerases (Koutsakos et al. 2019). PB1 is the most conserved protein across IAV and IBV, with about 60% amino acid identity. Vaccinating against the T cell epitope PB1413 might provide universal protection in humans. Besides, CD8+ T cell targets from IBV HA and NS1 proteins also have been identified and shown IBV-specific protection in mice, suggesting them as ideal targets for IBV-wide influenza vaccines (Koutsakos et al. 2019).
CD4+ T cell responses to influenza virus and their role for host protection against influenza infection have received increasing attention (Wilkinson et al. 2012). It has been well demonstrated that CD4+ memory T cells can help in directing a faster antibody response via cytokine secretion in response to mutated or immunologically novel viral antigens, leading to limited virus shedding and disease severity, possibly due to the direct lysis of infected epithelial cells (Poon et al. 2009; Wilkinson et al. 2012; Valkenburg et al. 2018). A vaccine that can induce CD8+ or CD4+ T responses against highly conserved regions of the influenza proteins may overcome the limitations of current season influenza vaccines (Sheikh et al. 2016). More importantly, universal T cell-inducing vaccines in combination with universal antibodies can confer increased protection against influenza (Asthagiri Arunkumar et al. 2019).
Obstacles and Strategies
In theory, a universal influenza vaccine would work well if it contains a conserved epitope that remains the same from year to year and does not vary between strains. However, no such viral epitope capable of stimulating an immune response that stops most influenza viruses afflicting humans has been reported yet (Cohen 2018). Certainly there are main obstacles on the way to developing universal influenza vaccines, but different strategies can be used to overcome them.
Low Immunogenicity and Immunofocusing
Specific induction of broadly neutralizing antibodies can be achieved by targeting B cell recognition to conserved epitopes. However, the low immunogenicity of the conserved HA stalk and M2e makes antibody responses targeting these antigens constitute only a small fraction of the total anti-virus antibodies in nature infections (Sui et al. 2011). Various immunofocusing approaches have been therefore developed to overcome this problem.
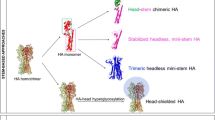
The M2e is small in size and has low abundance in its native state, but its immunogenicity can be improved using tandem repeats and fusing them to highly immunogenic carriers (Neirynck et al. 1999; De Filette et al. 2008; Eliasson et al. 2008; Turley et al. 2011; Kim et al. 2013). Strategies directing the antibody response to HA stalk were also evaluated, such as shielding of the HA head epitopes by hyperglycosylation (Fig. 3A) (Lin et al. 2012; Eggink et al. 2014), and constructing headless mini-HA as protective immunogen by removing the head domain completely (Fig. 3B) (Steel et al. 2010). Notably, the latter strategy requires maintaining the conformation of the stalk domain in absence of the head domain, which is challenging. Nevertheless promise progress has been made to achieve the stable native structure of trimeric HA stalk, either in soluble form or presented on nanoparticles (Lu et al. 2014; Mallajosyula et al. 2014; Impagliazzo et al. 2015; Yassine et al. 2015).
Immunofocusing approaches for a universal flu vaccine. A Head masking. The “Head” region of trimeric HA was hyperglycosylated, leaving only the HA stalk immunogenic. B Mini-HA. The “Head” region of trimeric HA was removed completely, while the conformation of the stalk domain maintains well. C Schematic of the PMD strategy. First, the epitope of interest is protected by binding of specific mAb. Then the surface of the protein·mAb complex is modified and rendered nonimmunogenic (shown as grey). By removal of the mAb, the epitope is deprotected and exposed as PMD antigen.
More recently, Weidenbacher and Kim developed a novel method, called protect, modify, deprotect (PMD), for creating immunogens aimed to elicit antibodies targeting a specific epitope (Weidenbacher and Kim 2019). A monoclonal antibody that recognizes the specific epitope is used to protect the target epitope on the protein, followed by modifications of the remaining exposed surfaces of the protein to render them nonimmunogenic. After removal of the monoclonal antibody, the epitope is deprotected and the resultant protein is modified at the surface other than the target epitope (Fig. 3C). The increasing numbers of broadly-neutralizing mAbs and corresponding conserved epitopes may provide attractive targets for the application of PMD in universal influenza vaccine design.
Immunodominance and Immunosubversion
The success of mini-HA in inducing robust stalk-specific antibodies strongly implies that the HA stalk is not intrinsically poorly immunogenic, but is the victim of immunodominance, where the immune system tends to respond to complex antigens in a reproducibly hierarchical manner, that is, higher-ranking antigens sometimes suppress responses to lower ranking antigens (Angeletti and Yewdell 2018). In the case of IAV HA, there is clear head immunodomination over stalk.
However, immunodominance can be subverted by various strategies. For example, an “antigen imprinting” strategy was inspired by the phenomenon that natural infection and vaccination with pandemic H1N1 in 2009 preferentially boosted broadly binding antibodies in humans including antibodies targeting the stalk domain (Wrammert et al. 2011; Li et al. 2012; Pica et al. 2012). Generally, immune response against newly emerging strains of influenza virus would be affected by immune memory acquired by past influenza exposure; memory B cells that are cross-reactive with newly emerging strains is activated, while induction of strain-specific antibodies is prevented (Henry et al. 2018). In this strategy, chimeric HAs that share the same stalk domain but express different head domains were constructed and used for vaccination sequentially, allowing repeated exposure of the same stalk in the context of an irrelevant globular head domain (Fig. 4A) (Hai et al. 2012; Krammer et al. 2013; Margine et al. 2013; Ermler et al. 2017). The first immunization with a chimeric HA construct leads to a primary response against the globular head domain and, though much less efficiently, against the stalk domain. Upon subsequent booster vaccinations, a recall response against the stalk is anticipated, but immune response against the immunodominant head domain is restricted.
immunosubversion strategies for a universal flu vaccine. A Chimeric hemagglutinin (cHA) constructs and vaccination strategy. The cHAs consist of a conserved stalk domain in combination with “heads” from different avian influenza virus HA subtypes. Vaccination with the first cHA leads to primary antibody responses mainly targeting the immunodominant head domain and low-level priming against the stalk domain. Upon boosting with a second cHA that has the same stalk domain but a completely different head domain, the immune system induces a primary response against the novel head domain and a strong memory response against the stalk domain. B Mosaic hemagglutinin (mHA) constructs and vaccination strategy. Only the variable immunodominant antigenic sites in mHAs are replaced with antigenic site equivalents from different influenza virus HA subtypes, and the resulting mHA displays conserved epitopes in both the stalk and head domains. Vaccination with the first mHA likely induces a strong primary response against the grafted antigenic sites and low-level priming for conserved epitopes in the stalk and head domains. Revaccination with a second mHA with antigenic sites that have been grafted from a different subtype might result in a primary response against the antigenic sites and a strong recall response against conserved epitopes in both the head and stalk domains. C Model of immunosubversion approach with mosaic antigen array. Colored and black simbols indicates strain specific and conserved antigens respectivley. B cells possessing BCRs specific to multiple antigenic variants are colored accordingly. The avidity advantage of the mosaic antigen to cross-reactive B cells over mono-specific B cells is anticipated, promoting proliferation of cross-reactive B cells.
To further include head-specific cross-reactive epitopes into consideration for universal vaccine design, Krammer and Palese proposed a novel concept of mosaic HAs, which were constructed by replacing the immunodominant antigenic sites in the head with sequences from exotic, avian HA subtypes, instead of changing the whole head domain (Fig. 4B) (Broecker et al. 2018; Liu et al. 2018; Krammer and Palese 2019). Such a vaccination regimen can induce antibodies targeting both conserved HA stalk and conserved epitopes in the head domain, possessing improved potential as universal vaccines (Sun et al. 2019).
Recently, Kanekiyo et al. developed a novel immunosubversion strategy by enhancing the avidity of immunogens to cross-reactive B cells (Fig. 4C) (Kanekiyo et al. 2019). Heterotypic influenza hemagglutinin antigens were co-localized on a single nanoparticle, generating a mosaic array. After vaccination, the avidity advantage of conserved epitopes to B cells can be achieved over strain-specific B cells, i.e., the B cell responses can adaptively target conserved antigenic surfaces, promoting cross-reactive antibody secretion. This heterotypic mosaic nanoparticle immunogen provides a new tool to combat viral genetic plasticity and antigenic variation.
Of note, a latest study using an African green monkey IAV model revealed that although mismatched prime-boost generated a pool of stem-specific memory B cells, head-specific B cells and serum Abs still substantially dominated the immune response (Jegaskanda et al. 2019). Moreover, it’s theoretically proven that, at least in prime-boost model of mice, memory B cell (MBC) clones seldom reenter secondary germinal centers upon boosting (Mesin et al. 2020). The rearrangement of MBC clones were therefore blocked, and the diversity and specificity were limited, restricting the breadth and effectiveness of the ensuing antibody response (Mesin et al. 2020). If this clonality bottleneck is also the case of humans, our ability to elicit antibodies to non-immunodominant epitopes may be further improved once the bottleneck is solved in future.
Original Antigenic Sin Versus Additive Approach
“Original Antigenic Sin (OAS)” refers to the phenomenon that the antibody response to the first influenza virus variant one encounters dominates the anti-influenza virus antibody response lifelong (reviewed in (Henry et al. 2018)). To date, the knowledge of how past influenza exposure shapes the response to subsequent antigenically distinct influenza strains remains obscure. Although “antigen imprinting” strategy has shown positive impact in the context of the development of a “stalk-based” universal influenza virus vaccine, the complex immune response underlying OAS more or less stymies the development of universal influenza vaccines (Cohen 2018).
Additive approach by either serial vaccination or combined vaccination with multiple antigenic types has been successfully employed to produce pneumococcal conjugate vaccines (13 different antigens) and human papillomavirus vaccines (up to nine different antigens), which can elicit accumulated responses with non-overlapping specificities. For influenza vaccines, admixture of multivalent influenza vaccines may result in antigenic competition and eventually lead to loss of efficacy for one or more components (Kanekiyo et al. 2019). Moreover, broad immunity has not been achieved by serial immunization with conventional flu vaccines (Belongia et al. 2017).
It is argued by Worobey et al. that the propensity of initial influenza virus exposure to establish a lifelong immunological imprint also presents a remarkable opportunity: immunization of infants prior to their initial, natural virus exposure with multiple versions of common human subtypes simultaneously may promise extended immunological imprinting across all currently circulating strains and against potential pandemic strains of IAV. This approach might be a possible step toward a universal vaccine (Cohen 2018; Worobey et al. 2020).
Future Prospects
Decades have been spent to concoct universal influenza vaccines, and some solid progress has been achieved. Currently, several vaccine candidates are in early human clinical trials, with each vaccine candidate exploiting conserved regions of the virus to maximize breadth [reviewed in (Nachbagauer and Krammer, 2017)]. Although some vaccine candidates such as those featured HA stalk have underwhelmed vaccine developers due to their poor effectiveness (Cohen 2018), increasing numbers of new effective broad-protective immunogens (epitopes) have been discovered, providing attractive targets that should be explored in the development of novel universal influenza vaccines (Bajic et al. 2019; Bangaru et al. 2019; Stadlbauer et al. 2019; Watanabe et al. 2019).
Of particularly note, some of the novel conserved epitopes are discontinuous (Stadlbauer et al. 2019) or even occluded in their native state (Bajic et al. 2019; Bangaru et al. 2019; Watanabe et al. 2019). How one can obtain and immunize against these epitopes remains a big issue. The advantage of synthetic epitope mimetics and de novo protein design may facilitate immunogens design for universal influenza vaccines in the future (Zerbe et al. 2017; Grayson and Anderson 2018).
Synthetic Epitope Mimetics
The huge growth in high-resolution 3D structures of proteins promotes the emergence of a novel paradigm in the study of biomolecular recognition, the design and synthesis of protein epitope mimetics. Based on the natural structure of interested epitope, the mimetics (synthetic peptides) can be designed and synthesized, with diverse chemistries and cross-links (e.g., helix-stabilizing staples) harnessed to achieve desired folding (Zerbe et al. 2017).
Recent technological advances have suggested the promising potential of synthetic peptides as antigens to generate focused immune responses. By coupling a mimetic of the V3 loop from HIV-1 GP120 to synthetic virus-like nanoparticles, Riedel et al. successfully designed a candidate HIV-1 vaccine which can elicit antibodies in rabbits that recognized recombinant gp120 (Riedel et al. 2011). Moreover, both folded B- and linear T- epitopes can be displayed on the engineered nanoparticles, inducing antibody response and T-cell response respectively (Zerbe et al. 2017).
De Novo Immunogen Design
The computational design of new proteins sparks hopes in the field of rational vaccinology, particularly to elicit targeted neutralizing antibody responses (Correia et al. 2014; Sesterhenn et al. 2019). While most de novo proteins designed so far are either functionless or present functions that are encoded by regular, continuous secondary structures. Interestingly, a great progress has been made recently by Sesterhenn et al., who developed a novel computational design strategy to build de novo proteins presenting complex structural motifs (Sesterhenn et al. 2020). By using this strategy, the authors further engineered epitope-focused immunogens mimicking irregular and discontinuous RSV neutralization epitopes. And excitingly, the de novo–designed immunogens can induce robust neutralizing responses against the respiratory syncytial virus in both mice and nonhuman primates (Sesterhenn et al. 2020).
Conclusions
The ever-changing influenza surface proteins, HA and NA, arouse the desire of a universal flu vaccine. By offering life-long protection against all influenza strains, an universal vaccine can greatly reduce the need for yearly vaccine reformulations and vaccination. So far, many universal influenza vaccine candidates that are in clinical trials focus on targeting conserved epitopes of influenza, including either surface proteins or highly conserved internal proteins. Moreover, the new identified highly conserved epitopes and advanced techniques will lead to the fast development of next-generation universal flu vaccines.
References
Angeletti D, Yewdell JW (2018) Is it possible to develop a “universal” influenza virus vaccine? Outflanking antibody immunodominance on the road to universal influenza vaccination. Cold Spring Harb Perspect Biol 10:a028852
Antrobus RD, Berthoud TK, Mullarkey CE, Hoschler K, Coughlan L, Zambon M, Hill AV, Gilbert SC (2014a) Coadministration of seasonal influenza vaccine and MVA-NP + M1 simultaneously achieves potent humoral and cell-mediated responses. Mol Ther 22:233–238
Antrobus RD, Coughlan L, Berthoud TK, Dicks MD, Hill AV, Lambe T, Gilbert SC (2014b) Clinical assessment of a novel recombinant simian adenovirus ChAdOx1 as a vectored vaccine expressing conserved Influenza A antigens. Mol Ther 22:668–674
Asthagiri Arunkumar G, McMahon M, Pavot V, Aramouni M, Ioannou A, Lambe T, Gilbert S, Krammer F (2019) Vaccination with viral vectors expressing NP, M1 and chimeric hemagglutinin induces broad protection against influenza virus challenge in mice. Vaccine 37:5567–5577
Bajic G, Maron MJ, Adachi Y, Onodera T, McCarthy KR, McGee CE, Sempowski GD, Takahashi Y, Kelsoe G, Kuraoka M, Schmidt AG (2019) Influenza antigen engineering focuses immune responses to a subdominant but broadly protective viral epitope. Cell Host Microb 25(827–835):e826
Bangaru S, Lang S, Schotsaert M, Vanderven HA, Zhu X, Kose N, Bombardi R, Finn JA, Kent SJ, Gilchuk P, Gilchuk I, Turner HL, Garcia-Sastre A, Li S, Ward AB, Wilson IA, Crowe JE Jr (2019) A site of vulnerability on the influenza virus hemagglutinin head domain trimer interface. Cell 177(1136–1152):e1118
Belongia EA, Skowronski DM, McLean HQ, Chambers C, Sundaram ME, De Serres G (2017) Repeated annual influenza vaccination and vaccine effectiveness: review of evidence. Expert Rev Vaccin 16:1–14
Berthoud TK, Hamill M, Lillie PJ, Hwenda L, Collins KA, Ewer KJ, Milicic A, Poyntz HC, Lambe T, Fletcher HA, Hill AV, Gilbert SC (2011) Potent CD8 + T-cell immunogenicity in humans of a novel heterosubtypic influenza A vaccine, MVA-NP + M1. Clin Infect Dis 52:1–7
Boonsathorn N, Panthong S, Koksunan S, Chittaganpitch M, Phuygun S, Waicharoen S, Prachasupap A, Sasaki T, Kubota-Koketsu R, Yasugi M, Ono K, Arai Y, Kurosu T, Sawanpanyalert P, Ikuta K, Watanabe Y (2014) A human monoclonal antibody derived from a vaccinated volunteer recognizes heterosubtypically a novel epitope on the hemagglutinin globular head of H1 and H9 influenza A viruses. Biochem Biophys Res Commun 452:865–870
Boyoglu-Barnum S, Hutchinson GB, Boyington JC, Moin SM, Gillespie RA, Tsybovsky Y, Stephens T, Vaile JR, Lederhofer J, Corbett KS, Fisher BE, Yassine HM, Andrews SF, Crank MC, McDermott AB, Mascola JR, Graham BS, Kanekiyo M (2020) Glycan repositioning of influenza hemagglutinin stem facilitates the elicitation of protective cross-group antibody responses. Nat Commun 11:791
Broecker F, Liu STH, Sun W, Krammer F, Simon V, Palese P (2018) Immunodominance of antigenic site B in the hemagglutinin of the current H3N2 influenza virus in humans and mice. J Virol 92:e01100-18
Bui HH, Peters B, Assarsson E, Mbawuike I, Sette A (2007) Ab and T cell epitopes of influenza A virus, knowledge and opportunities. Proc Natl Acad Sci U S A 104:246–251
Caton AJ, Brownlee GG, Yewdell JW, Gerhard W (1982) The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 31:417–427
Chen YQ, Wohlbold TJ, Zheng NY, Huang M, Huang Y, Neu KE, Lee J, Wan H, Rojas KT, Kirkpatrick E, Henry C, Palm AE, Stamper CT, Lan LY, Topham DJ, Treanor J, Wrammert J, Ahmed R, Eichelberger MC, Georgiou G, Krammer F, Wilson PC (2018) Influenza infection in humans induces broadly cross-reactive and protective neuraminidase-reactive antibodies. Cell 173:417–429.e10
Cohen J (2018) Universal flu vaccine is ‘an alchemist’s dream’. Science 362:1094
Correia BE, Bates JT, Loomis RJ, Baneyx G, Carrico C, Jardine JG, Rupert P, Correnti C, Kalyuzhniy O, Vittal V, Connell MJ, Stevens E, Schroeter A, Chen M, Macpherson S, Serra AM, Adachi Y, Holmes MA, Li Y, Klevit RE, Graham BS, Wyatt RT, Baker D, Strong RK, Crowe JE Jr, Johnson PR, Schief WR (2014) Proof of principle for epitope-focused vaccine design. Nature 507:201–206
Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, Silacci C, Fernandez-Rodriguez BM, Agatic G, Bianchi S, Giacchetto-Sasselli I, Calder L, Sallusto F, Collins P, Haire LF, Temperton N, Langedijk JP, Skehel JJ, Lanzavecchia A (2011) A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333:850–856
De Filette M, Martens W, Roose K, Deroo T, Vervalle F, Bentahir M, Vandekerckhove J, Fiers W, Saelens X (2008) An influenza A vaccine based on tetrameric ectodomain of matrix protein 2. J Biol Chem 283:11382–11387
de Jong JC, Beyer WE, Palache AM, Rimmelzwaan GF, Osterhaus AD (2000) Mismatch between the 1997/1998 influenza vaccine and the major epidemic A(H3N2) virus strain as the cause of an inadequate vaccine-induced antibody response to this strain in the elderly. J Med Virol 61:94–99
Deng L, Wang BZ (2018) A perspective on nanoparticle universal influenza vaccines. ACS Infect Dis 4:1656–1665
Deng L, Cho KJ, Fiers W, Saelens X (2015) M2e-based universal influenza A vaccines. Vaccin Basel 3:105–136
DiLillo DJ, Tan GS, Palese P, Ravetch JV (2014) Broadly neutralizing hemagglutinin stalk-specific antibodies require FcgammaR interactions for protection against influenza virus in vivo. Nat Med 20:143–151
Doyle TM, Hashem AM, Li C, Van Domselaar G, Larocque L, Wang J, Smith D, Cyr T, Farnsworth A, He R, Hurt AC, Brown EG, Li X (2013) Universal anti-neuraminidase antibody inhibiting all influenza A subtypes. Antivir Res 100:567–574
Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, van der Vlugt R, Lamrani M, Korse HJ, Geelen E, Sahin O, Sieuwerts M, Brakenhoff JP, Vogels R, Li OT, Poon LL, Peiris M, Koudstaal W, Ward AB, Wilson IA, Goudsmit J, Friesen RH (2012) Highly conserved protective epitopes on influenza B viruses. Science 337:1343–1348
Du R, Cui Q, Rong L (2019) Competitive cooperation of hemagglutinin and neuraminidase during influenza A virus entry. Viruses 11:458
Eggink D, Goff PH, Palese P (2014) Guiding the immune response against influenza virus hemagglutinin toward the conserved stalk domain by hyperglycosylation of the globular head domain. J Virol 88:699–704
Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA (2009) Antibody recognition of a highly conserved influenza virus epitope. Science 324:246–251
Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJ, Brandenburg B, Vogels R, Brakenhoff JP, Kompier R, Koldijk MH, Cornelissen LA, Poon LL, Peiris M, Koudstaal W, Wilson IA, Goudsmit J (2011) A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333:843–850
Ekiert DC, Kashyap AK, Steel J, Rubrum A, Bhabha G, Khayat R, Lee JH, Dillon MA, O’Neil RE, Faynboym AM, Horowitz M, Horowitz L, Ward AB, Palese P, Webby R, Lerner RA, Bhatt RR, Wilson IA (2012) Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 489:526–532
Eliasson DG, El Bakkouri K, Schon K, Ramne A, Festjens E, Lowenadler B, Fiers W, Saelens X, Lycke N (2008) CTA1-M2e-DD: a novel mucosal adjuvant targeted influenza vaccine. Vaccine 26:1243–1252
Epstein SL (2006) Prior H1N1 influenza infection and susceptibility of Cleveland Family Study participants during the H2N2 pandemic of 1957: an experiment of nature. J Infect Dis 193:49–53
Ermler ME, Kirkpatrick E, Sun W, Hai R, Amanat F, Chromikova V, Palese P, Krammer F (2017) Chimeric hemagglutinin constructs induce broad protection against influenza B virus challenge in the mouse model. J Virol 91:e00286-17
Farahmand B, Taheri N, Shokouhi H, Soleimanjahi H, Fotouhi F (2019) Chimeric protein consisting of 3M2e and HSP as a universal influenza vaccine candidate: from in silico analysis to preliminary evaluation. Virus Genes 55:22–32
Florek NW, Weinfurter JT, Jegaskanda S, Brewoo JN, Powell TD, Young GR, Das SC, Hatta M, Broman KW, Hungnes O, Dudman SG, Kawaoka Y, Kent SJ, Stinchcomb DT, Osorio JE, Friedrich TC (2014) Modified vaccinia virus Ankara encoding influenza virus hemagglutinin induces heterosubtypic immunity in macaques. J Virol 88:13418–13428
Gerdil C (2003) The annual production cycle for influenza vaccine. Vaccine 21:1776–1779
Gras S, Kedzierski L, Valkenburg SA, Laurie K, Liu YC, Denholm JT, Richards MJ, Rimmelzwaan GF, Kelso A, Doherty PC, Turner SJ, Rossjohn J, Kedzierska K (2010) Cross-reactive CD8 + T-cell immunity between the pandemic H1N1-2009 and H1N1-1918 influenza A viruses. Proc Natl Acad Sci U S A 107:12599–12604
Grayson KJ, Anderson JLR (2018) Designed for life: biocompatible de novo designed proteins and components. J R Soc Interface 15:20180472
Greenbaum JA, Kotturi MF, Kim Y, Oseroff C, Vaughan K, Salimi N, Vita R, Ponomarenko J, Scheuermann RH, Sette A, Peters B (2009) Pre-existing immunity against swine-origin H1N1 influenza viruses in the general human population. Proc Natl Acad Sci U S A 106:20365–20370
Hai R, Krammer F, Tan GS, Pica N, Eggink D, Maamary J, Margine I, Albrecht RA, Palese P (2012) Influenza viruses expressing chimeric hemagglutinins: globular head and stalk domains derived from different subtypes. J Virol 86:5774–5781
Hayward AC, Wang L, Goonetilleke N, Fragaszy EB, Bermingham A, Copas A, Dukes O, Millett ER, Nazareth I, Nguyen-Van-Tam JS, Watson JM, Zambon M, Johnson AM, McMichael AJ (2015) Natural T cell-mediated protection against seasonal and pandemic influenza. Results of the flu watch cohort study. Am J Respir Crit Care Med 191:1422–1431
He W, Mullarkey CE, Duty JA, Moran TM, Palese P, Miller MS (2015) Broadly neutralizing anti-influenza virus antibodies: enhancement of neutralizing potency in polyclonal mixtures and IgA backbones. J Virol 89:3610–3618
Heaton NS, Sachs D, Chen CJ, Hai R, Palese P (2013) Genome-wide mutagenesis of influenza virus reveals unique plasticity of the hemagglutinin and NS1 proteins. Proc Natl Acad Sci U S A 110:20248–20253
Henry C, Palm AE, Krammer F, Wilson PC (2018) From original antigenic sin to the universal influenza virus vaccine. Trends Immunol 39:70–79
Huang Y, Owino SO, Crevar CJ, Carter DM, Ross TM (2019) N-linked glycans and K147 residue on hemagglutinin synergize to elicit broadly-reactive H1N1 influenza antibodies. J Virol. https://doi.org/10.1128/JVI.01432-19
Impagliazzo A, Milder F, Kuipers H, Wagner MV, Zhu X, Hoffman RM, van Meersbergen R, Huizingh J, Wanningen P, Verspuij J, de Man M, Ding Z, Apetri A, Kukrer B, Sneekes-Vriese E, Tomkiewicz D, Laursen NS, Lee PS, Zakrzewska A, Dekking L, Tolboom J, Tettero L, van Meerten S, Yu W, Koudstaal W, Goudsmit J, Ward AB, Meijberg W, Wilson IA, Radosevic K (2015) A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 349:1301–1306
Jegaskanda S, Job ER, Kramski M, Laurie K, Isitman G, de Rose R, Winnall WR, Stratov I, Brooks AG, Reading PC, Kent SJ (2013a) Cross-reactive influenza-specific antibody-dependent cellular cytotoxicity antibodies in the absence of neutralizing antibodies. J Immunol 190:1837–1848
Jegaskanda S, Weinfurter JT, Friedrich TC, Kent SJ (2013b) Antibody-dependent cellular cytotoxicity is associated with control of pandemic H1N1 influenza virus infection of macaques. J Virol 87:5512–5522
Jegaskanda S, Reading PC, Kent SJ (2014) Influenza-specific antibody-dependent cellular cytotoxicity: toward a universal influenza vaccine. J Immunol 193:469–475
Jegaskanda S, Andrews SF, Wheatley AK, Yewdell JW, McDermott AB, Subbarao K (2019) Hemagglutinin head-specific responses dominate over stem-specific responses following prime boost with mismatched vaccines. JCI Insight 4:e129035
Kanekiyo M, Joyce MG, Gillespie RA, Gallagher JR, Andrews SF, Yassine HM, Wheatley AK, Fisher BE, Ambrozak DR, Creanga A, Leung K, Yang ES, Boyoglu-Barnum S, Georgiev IS, Tsybovsky Y, Prabhakaran MS, Andersen H, Kong W-P, Baxa U, Zephir KL, Ledgerwood JE, Koup RA, Kwong PD, Harris AK, McDermott AB, Mascola JR, Graham BS (2019) Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nat Immunol 20:362–372
Kim MC, Song JM, Eunju O, Kwon YM, Lee YJ, Compans RW, Kang SM (2013) Virus-like particles containing multiple M2 extracellular domains confer improved cross-protection against various subtypes of influenza virus. Mol Ther 21:485–492
Koutsakos M, Wheatley AK, Loh L, Clemens EB, Sant S, Nussing S, Fox A, Chung AW, Laurie KL, Hurt AC, Rockman S, Lappas M, Loudovaris T, Mannering SI, Westall GP, Elliot M, Tangye SG, Wakim LM, Kent SJ, Nguyen THO, Kedzierska K (2018) Circulating TFH cells, serological memory, and tissue compartmentalization shape human influenza-specific B cell immunity. Sci Transl Med 10:eaan8405
Koutsakos M, Illing PT, Nguyen THO, Mifsud NA, Crawford JC, Rizzetto S, Eltahla AA, Clemens EB, Sant S, Chua BY, Wong CY, Allen EK, Teng D, Dash P, Boyd DF, Grzelak L, Zeng W, Hurt AC, Barr I, Rockman S, Jackson DC, Kotsimbos TC, Cheng AC, Richards M, Westall GP, Loudovaris T, Mannering SI, Elliott M, Tangye SG, Wakim LM, Rossjohn J, Vijaykrishna D, Luciani F, Thomas PG, Gras S, Purcell AW, Kedzierska K (2019) Human CD8(+) T cell cross-reactivity across influenza A, B and C viruses. Nat Immunol 20:613–625
Krammer F (2019) The human antibody response to influenza A virus infection and vaccination. Nat Rev Immunol 19:383–397
Krammer F, Palese P (2015) Advances in the development of influenza virus vaccines. Nat Rev Drug Discov 14:167–182
Krammer F, Palese P (2019) Universal influenza virus vaccines that target the conserved hemagglutinin stalk and conserved sites in the head domain. J Infect Dis 219:S62–S67
Krammer F, Pica N, Hai R, Margine I, Palese P (2013) Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J Virol 87:6542–6550
Krammer F, Smith GJD, Fouchier RAM, Peiris M, Kedzierska K, Doherty PC, Palese P, Shaw ML, Treanor J, Webster RG, Garcia-Sastre A (2018) Influenza. Nat Rev Dis Primers 4:3
Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Albrecht R, Blum DL, Ramos I, Fernandez-Sesma A, Edwards KM, Garcia-Sastre A, Basler CF, Crowe JE Jr (2012) Human monoclonal antibodies to pandemic 1957 H2N2 and pandemic 1968 H3N2 influenza viruses. J Virol 86:6334–6340
Lambe T, Carey JB, Li Y, Spencer AJ, van Laarhoven A, Mullarkey CE, Vrdoljak A, Moore AC, Gilbert SC (2013) Immunity against heterosubtypic influenza virus induced by adenovirus and MVA expressing nucleoprotein and matrix protein-1. Sci Rep 3:1443
Lee PS, Yoshida R, Ekiert DC, Sakai N, Suzuki Y, Takada A, Wilson IA (2012) Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity. Proc Natl Acad Sci U S A 109:17040–17045
Lee PS, Ohshima N, Stanfield RL, Yu W, Iba Y, Okuno Y, Kurosawa Y, Wilson IA (2014) Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus. Nat Commun 5:3614
Li GM, Chiu C, Wrammert J, McCausland M, Andrews SF, Zheng NY, Lee JH, Huang M, Qu X, Edupuganti S, Mulligan M, Das SR, Yewdell JW, Mehta AK, Wilson PC, Ahmed R (2012) Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc Natl Acad Sci U S A 109:9047–9052
Lillie PJ, Berthoud TK, Powell TJ, Lambe T, Mullarkey C, Spencer AJ, Hamill M, Peng Y, Blais ME, Duncan CJ, Sheehy SH, Havelock T, Faust SN, Williams RL, Gilbert A, Oxford J, Dong T, Hill AV, Gilbert SC (2012) Preliminary assessment of the efficacy of a T-cell-based influenza vaccine, MVA-NP + M1, in humans. Clin Infect Dis 55:19–25
Lin SC, Lin YF, Chong P, Wu SC (2012) Broader neutralizing antibodies against H5N1 viruses using prime-boost immunization of hyperglycosylated hemagglutinin DNA and virus-like particles. PLoS ONE 7:e39075
Liu W, Li H, Chen YH (2003) N-terminus of M2 protein could induce antibodies with inhibitory activity against influenza virus replication. FEMS Immunol Med Microbiol 35:141–146
Liu STH, Behzadi MA, Sun W, Freyn AW, Liu WC, Broecker F, Albrecht RA, Bouvier NM, Simon V, Nachbagauer R, Krammer F, Palese P (2018) Antigenic sites in influenza H1 hemagglutinin display species-specific immunodominance. J Clin Invest 128:4992–4996
Lu Y, Welsh JP, Swartz JR (2014) Production and stabilization of the trimeric influenza hemagglutinin stem domain for potentially broadly protective influenza vaccines. Proc Natl Acad Sci U S A 111:125–130
Mallajosyula VV, Citron M, Ferrara F, Lu X, Callahan C, Heidecker GJ, Sarma SP, Flynn JA, Temperton NJ, Liang X, Varadarajan R (2014) Influenza hemagglutinin stem-fragment immunogen elicits broadly neutralizing antibodies and confers heterologous protection. Proc Natl Acad Sci U S A 111:E2514–E2523
Mandala VS, Liao SY, Gelenter MD, Hong M (2019) The Transmembrane conformation of the influenza B virus M2 protein in lipid bilayers. Sci Rep 9:3725
Margine I, Krammer F, Hai R, Heaton NS, Tan GS, Andrews SA, Runstadler JA, Wilson PC, Albrecht RA, Garcia-Sastre A, Palese P (2013) Hemagglutinin stalk-based universal vaccine constructs protect against group 2 influenza A viruses. J Virol 87:10435–10446
McMichael AJ, Gotch FM, Noble GR, Beare PA (1983) Cytotoxic T-cell immunity to influenza. N Engl J Med 309:13–17
Memoli MJ, Han A, Walters KA, Czajkowski L, Reed S, Athota R, Rosas LA, Cervantes-Medina A, Park JK, Morens DM, Kash JC, Taubenberger JK (2019) Influenza A reinfection in sequential human challenge: implications for protective immunity and “universal” vaccine development. Clin Infect Dis. https://doi.org/10.1093/cid/ciz281
Mesin L, Schiepers A, Ersching J, Barbulescu A, Cavazzoni CB, Angelini A, Okada T, Kurosaki T, Victora GD (2020) Restricted clonality and limited germinal center reentry characterize memory B cell reactivation by boosting. Cell 180:92–106.e11
Mezhenskaya D, Isakova-Sivak I, Rudenko L (2019) M2e-based universal influenza vaccines: a historical overview and new approaches to development. J Biomed Sci 26:76
Miller MS, Tsibane T, Krammer F, Hai R, Rahmat S, Basler CF, Palese P (2013) 1976 and 2009 H1N1 influenza virus vaccines boost anti-hemagglutinin stalk antibodies in humans. J Infect Dis 207:98–105
Monto AS, Kendal AP (1973) Effect of neuraminidase antibody on Hong Kong influenza. Lancet 1:623–625
Murphy BR, Kasel JA, Chanock RM (1972) Association of serum anti-neuraminidase antibody with resistance to influenza in man. N Engl J Med 286:1329–1332
Nachbagauer R, Krammer F (2017) Universal influenza virus vaccines and therapeutic antibodies. Clin Microbiol Infect 23:222–228
Neirynck S, Deroo T, Saelens X, Vanlandschoot P, Jou WM, Fiers W (1999) A universal influenza A vaccine based on the extracellular domain of the M2 protein. Nat Med 5:1157–1163
Okuno Y, Isegawa Y, Sasao F, Ueda S (1993) A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol 67:2552–2558
Pica N, Hai R, Krammer F, Wang TT, Maamary J, Eggink D, Tan GS, Krause JC, Moran T, Stein CR, Banach D, Wrammert J, Belshe RB, Garcia-Sastre A, Palese P (2012) Hemagglutinin stalk antibodies elicited by the 2009 pandemic influenza virus as a mechanism for the extinction of seasonal H1N1 viruses. Proc Natl Acad Sci U S A 109:2573–2578
Poon LLM, Leung YHC, Nicholls JM, Perera P-Y, Lichy JH, Yamamoto M, Waldmann TA, Peiris JSM, Perera LP (2009) Vaccinia virus-based multivalent H5N1 avian influenza vaccines adjuvanted with IL-15 confer sterile cross-clade protection in mice. J Immunol Baltim Md 1950 182:3063–3071
Quinones-Parra S, Grant E, Loh L, Nguyen TH, Campbell KA, Tong SY, Miller A, Doherty PC, Vijaykrishna D, Rossjohn J, Gras S, Kedzierska K (2014) Preexisting CD8 + T-cell immunity to the H7N9 influenza A virus varies across ethnicities. Proc Natl Acad Sci U S A 111:1049–1054
Riedel T, Ghasparian A, Moehle K, Rusert P, Trkola A, Robinson JA (2011) Synthetic virus-like particles and conformationally constrained peptidomimetics in vaccine design. ChemBioChem 12:2829–2836
Schulman JL, Khakpour M, Kilbourne ED (1968) Protective effects of specific immunity to viral neuraminidase on influenza virus infection of mice. J Virol 2:778–786
Sesterhenn F, Galloux M, Vollers SS, Csepregi L, Yang C, Descamps D, Bonet J, Friedensohn S, Gainza P, Corthésy P, Chen M, Rosset S, Rameix-Welti MA, Éléouët JF, Reddy ST, Graham BS, Riffault S, Correia BE (2019) Boosting subdominant neutralizing antibody responses with a computationally designed epitope-focused immunogen. PLoS Biol 17:e3000164
Sesterhenn F, Yang C, Bonet J, Cramer JT, Wen X, Wang Y, Chiang CI, Abriata LA, Kucharska I, Castoro G, Vollers SS, Galloux M, Dheilly E, Rosset S, Corthésy P, Georgeon S, Villard M, Richard CA, Descamps D, Delgado T, Oricchio E, Rameix-Welti MA, Más V, Ervin S, Eléouët JF, Riffault S, Bates JT, Julien JP, Li Y, Jardetzky T, Krey T, Correia BE (2020) De novo protein design enables the precise induction of RSV-neutralizing antibodies. Science 368:eaay5051
Sheikh QM, Gatherer D, Reche PA, Flower DR (2016) Towards the knowledge-based design of universal influenza epitope ensemble vaccines. Bioinform Oxf Engl 32:3233–3239
Sridhar S, Begom S, Bermingham A, Hoschler K, Adamson W, Carman W, Bean T, Barclay W, Deeks JJ, Lalvani A (2013) Cellular immune correlates of protection against symptomatic pandemic influenza. Nat Med 19:1305–1312
Stadlbauer D, Zhu X, McMahon M, Turner JS, Wohlbold TJ, Schmitz AJ, Strohmeier S, Yu W, Nachbagauer R, Mudd PA, Wilson IA, Ellebedy AH, Krammer F (2019) Broadly protective human antibodies that target the active site of influenza virus neuraminidase. Science 366:499–504
Steel J, Lowen AC, Wang TT, Yondola M, Gao Q, Haye K, Garcia-Sastre A, Palese P (2010) Influenza virus vaccine based on the conserved hemagglutinin stalk domain. MBio 1:e00018-10
Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, Wan H, Murakami A, Yammanuru A, Han T, Cox NJ, Bankston LA, Donis RO, Liddington RC, Marasco WA (2009) Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol 16:265–273
Sui J, Sheehan J, Hwang WC, Bankston LA, Burchett SK, Huang CY, Liddington RC, Beigel JH, Marasco WA (2011) Wide prevalence of heterosubtypic broadly neutralizing human anti-influenza A antibodies. Clin Infect Dis 52:1003–1009
Sun W, Kirkpatrick E, Ermler M, Nachbagauer R, Broecker F, Krammer F, Palese P (2019) Development of influenza B universal vaccine candidates using the “Mosaic” hemagglutinin approach. J Virol 93:e00333-19
Terajima M, Cruz J, Co MD, Lee JH, Kaur K, Wrammert J, Wilson PC, Ennis FA (2011) Complement-dependent lysis of influenza a virus-infected cells by broadly cross-reactive human monoclonal antibodies. J Virol 85:13463–13467
Tobler K, Kelly ML, Pinto LH, Lamb RA (1999) Effect of cytoplasmic tail truncations on the activity of the M(2) ion channel of influenza A virus. J Virol 73:9695–9701
Treanor JJ, Tierney EL, Zebedee SL, Lamb RA, Murphy BR (1990) Passively transferred monoclonal antibody to the M2 protein inhibits influenza A virus replication in mice. J Virol 64:1375–1377
Turley CB, Rupp RE, Johnson C, Taylor DN, Wolfson J, Tussey L, Kavita U, Stanberry L, Shaw A (2011) Safety and immunogenicity of a recombinant M2e-flagellin influenza vaccine (STF2.4xM2e) in healthy adults. Vaccine 29:5145–5152
Valkenburg SA, Li OTW, Li A, Bull M, Waldmann TA, Perera LP, Peiris M, Poon LLM (2018) Protection by universal influenza vaccine is mediated by memory CD4 T cells. Vaccine 36:4198–4206
Wan H, Gao J, Xu K, Chen H, Couzens LK, Rivers KH, Easterbrook JD, Yang K, Zhong L, Rajabi M, Ye J, Sultana I, Wan XF, Liu X, Perez DR, Taubenberger JK, Eichelberger MC (2013) Molecular basis for broad neuraminidase immunity: conserved epitopes in seasonal and pandemic H1N1 as well as H5N1 influenza viruses. J Virol 87:9290–9300
Wang TT, Palese P (2011) Biochemistry. Catching a moving target. Science 333:834–835
Wang Z, Wan Y, Qiu C, Quinones-Parra S, Zhu Z, Loh L, Tian D, Ren Y, Hu Y, Zhang X, Thomas PG, Inouye M, Doherty PC, Kedzierska K, Xu J (2015) Recovery from severe H7N9 disease is associated with diverse response mechanisms dominated by CD8(+) T cells. Nat Commun 6:6833
Wang Z, Zhu L, Nguyen THO, Wan Y, Sant S, Quinones-Parra SM, Crawford JC, Eltahla AA, Rizzetto S, Bull RA, Qiu C, Koutsakos M, Clemens EB, Loh L, Chen T, Liu L, Cao P, Ren Y, Kedzierski L, Kotsimbos T, McCaw JM, La Gruta NL, Turner SJ, Cheng AC, Luciani F, Zhang X, Doherty PC, Thomas PG, Xu J, Kedzierska K (2018) Clonally diverse CD38(+)HLA-DR(+)CD8(+) T cells persist during fatal H7N9 disease. Nat Commun 9:824
Wanitchang A, Wongthida P, Jongkaewwattana A (2016) Influenza B virus M2 protein can functionally replace its influenza A virus counterpart in promoting virus replication. Virology 498:99–108
Watanabe A, McCarthy KR, Kuraoka M, Schmidt AG, Adachi Y, Onodera T, Tonouchi K, Caradonna TM, Bajic G, Song S, McGee CE, Sempowski GD, Feng F, Urick P, Kepler TB, Takahashi Y, Harrison SC, Kelsoe G (2019) Antibodies to a conserved influenza head interface epitope protect by an IgG subtype-dependent mechanism. Cell 177(1124–1135):e1116
Weidenbacher PA, Kim PS (2019) Protect, modify, deprotect (PMD): a strategy for creating vaccines to elicit antibodies targeting a specific epitope. Proc Natl Acad Sci U S A 116:9947–9952
Wilkinson TM, Li CKF, Chui CSC, Huang AKY, Perkins M, Liebner JC, Lambkin-Williams R, Gilbert A, Oxford J, Nicholas B, Staples KJ, Dong T, Douek DC, McMichael AJ, Xu X-N (2012) Preexisting influenza-specific CD4 + T cells correlate with disease protection against influenza challenge in humans. Nat Med 18:274–280
Wilson JR, Guo Z, Reber A, Kamal RP, Music N, Gansebom S, Bai Y, Levine M, Carney P, Tzeng WP, Stevens J, York IA (2016) An influenza A virus (H7N9) anti-neuraminidase monoclonal antibody with prophylactic and therapeutic activity in vivo. Antivir Res 135:48–55
Wohlbold TJ, Podolsky KA, Chromikova V, Kirkpatrick E, Falconieri V, Meade P, Amanat F, Tan J, tenOever BR, Tan GS, Subramaniam S, Palese P, Krammer F (2017) Broadly protective murine monoclonal antibodies against influenza B virus target highly conserved neuraminidase epitopes. Nat Microbiol 2:1415–1424
World Health Organization (WHO) (2018) 2018 Influenza (seasonal) fact sheet. WHO, Geneva, Switzerland. http://www.who.int/mediacentre/factsheets/fs211/en/. Accessed 6 June 2020
Worobey M, Plotkin S, Hensley SE (2020) Influenza vaccines delivered in early childhood could turn antigenic sin into antigenic blessings. Cold Spring Harb Perspect Med. https://doi.org/10.1101/cshperspect.a038471
Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M, McCausland M, Skountzou I, Hornig M, Lipkin WI, Mehta A, Razavi B, Del Rio C, Zheng NY, Lee JH, Huang M, Ali Z, Kaur K, Andrews S, Amara RR, Wang Y, Das SR, O’Donnell CD, Yewdell JW, Subbarao K, Marasco WA, Mulligan MJ, Compans R, Ahmed R, Wilson PC (2011) Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med 208:181–193
Wu Y, Gao GF (2019) “Breathing” hemagglutinin reveals cryptic epitopes for universal influenza vaccine design. Cell 177:1086–1088
Wu NC, Wilson IA (2018) Structural insights into the design of novel anti-influenza therapies. Nat Struct Mol Biol 25:115–121
Yao Y, Wang H, Chen J, Shao Z, He B, Lan J, Chen Q, Chen Z (2019) Protection against homo and hetero-subtypic in fluenza A virus by optimized M2e DNA vaccine. Emerg Microb Infect 8:45–54
Yassine HM, Boyington JC, McTamney PM, Wei CJ, Kanekiyo M, Kong WP, Gallagher JR, Wang L, Zhang Y, Joyce MG, Lingwood D, Moin SM, Andersen H, Okuno Y, Rao SS, Harris AK, Kwong PD, Mascola JR, Nabel GJ, Graham BS (2015) Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med 21:1065–1070
Zebedee SL, Lamb RA (1989) Growth restriction of influenza A virus by M2 protein antibody is genetically linked to the M1 protein. Proc Natl Acad Sci U S A 86:1061–1065
Zerbe K, Moehle K, Robinson JA (2017) Protein epitope mimetics: from new antibiotics to supramolecular synthetic vaccines. Acc Chem Res 50:1323–1331
Acknowledgements
This work was supported by The Drug Innovation Major Project (Grant No. 2018ZX09711001), the Key Research and Development Projects of Science and Technology Department of Shandong Province (Grant No. 2017CXGC1309) and Shandong Provincial Natural Science Foundation of China (Grant No. ZR2019MH078; ZR2017MH086).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Animal and Human Rights Statement
This article does not contain any studies with human or animal subjects performed by any of the authors.
Rights and permissions
About this article
Cite this article
Du, R., Cui, Q. & Rong, L. Flu Universal Vaccines: New Tricks on an Old Virus. Virol. Sin. 36, 13–24 (2021). https://doi.org/10.1007/s12250-020-00283-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12250-020-00283-6