Abstract
The occurrence and antibiotic susceptibility profile of Staphylococcus isolates of healthy farm animal origin in Nkonkobe Municipality as well as the prevalence of putative antibiotic resistance genes were investigated using phenotypic and molecular methods. A total of 120 Staphylococcus isolates were isolated from 150 animal samples and consisted of Staphylococcus haemolyticus (30 %) and Staphylococcus aureus (23.3 %) from pig, Staphylococcus capitis (15 %) from goat, S. haemolyticus (5 %) and Staphylococcus xylosus (15 %) from cattle, and other staphylococci (11.7 %) from dead chicken and pigs. Besides this, the presence of these isolates was observed from the animal dung, showing that the organisms are shed to the environment. About 23.3 % of these isolates were coagulase-positive and 76.7 % were coagulase-negative Staphylococcus. Between 75 and 100 % of the isolates were resistant to penicillin G, tetracycline, sulfamethoxazole, and nalidixic acid; about 38 % were methicillin-resistant staphylococci, including 12.6 % methicillin-resistant S. aureus from pigs. In total, 12 % of all isolates were vancomycin resistant. Also, 12 % of the isolates were erythromycin resistant, while 40.2 % were resistant to ceftazidime. Only the genes mecA and mphC could be confirmed, whereas the genes vanA, vanB, ermA, ermB, and ermC could not be detected. The high phenotypic antibiotic resistance and the presence of some associated resistance genes is a potential threat to public health and suggest the animals to be important reservoirs of antibiotic resistance determinants in the environment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The role of commensals in the spread of resistance among bacteria around the globe cannot be overemphasized (Levy 1986; Summers 2002). They have been fingered as the possible sources of antibiotic resistance genes which are now rampant among many systemic bacterial pathogens of suppurative and non-suppurative infections (Saha et al. 2007; Kitara et al. 2011). The prior exposure of the commensals to antibiotics during chemotherapy for other infections in their hosts might be a selective force for antibiotic resistance (Cohen 1992), especially in human and in animal husbandry, where antibiotics are used in large quantities (Moulin 2001; Harada and Asai 2010). A number of glycopeptides being used as growth promoters in animal husbandry have been reported as inducers of antibiotic resistance in the animal body flora (Perrier-Gros-Claude et al. 1998; Hammerum et al. 2010). The subsequent emergence of resistance has heightened bacterial colonization and/or invasion of the animals, with reduced response to control leading to their persistence as contaminants in animal products such as unpasteurized milk (Kaur and Pathania 2010; Ogbodo et al. 2011). Zoonotic transfer of such difficult-to-treat bacterial species is becoming a worrisome trend, as it increases the disease burden in sub-Saharan African countries, leads to higher cost of procuring treatment options, and increases infant mortality (WHO 2011)
Staphylococcus spp. are among the important commensals of farm animals that often bear resistance genes. It is worthy to recall that the emergence of methicillin resistance was reported in S. aureus following the antibiotic introduction in the 1960s (Grundmann et al. 2006). This attribute was first reported in hospitals but later in the community. Today, the methicillin-resistant S. aureus (MRSA) is still a threat to global health and wellness (Van Loo et al. 2007). Other previously known commensal Staphylococcus spp. now constitute threats to global health. Some coagulase-negative staphylococci (CoNS) that colonize animal skin and mucous membrane as flora (Kloos and Bannerman 1994) are now implicated in skin and soft tissue infection of man (Shittu et al. 2004), bacteriemia, and septicemia (Komolafe and Adegoke 2008; D'mello et al. 2008). Staphylococcus haemolyticus, Staphylococcus capitis, and Staphylococcus xylosus which are commensals of farm animals can also cause opportunistic infection(s) in other cases (Thorberg 2008).
Weese et al. (2005) and Walther et al. (2008) reported the notable presence of MRSA in animals. The MRSA has been increasingly reported in cattle, goat, sheep, cats, camel, and dogs, and have been examined for resistance to such vital antibiotics by Walther et al. (2008) and Saleha et al. (2010). While other animals also harbor MRSA alike, pigs are vital examples, known to harbor typical MRSA which spreads rapidly among the entire swine (Cuny et al. 2010; Van den Broek et al. 2009). These strains have been found among piggery workers, suggesting a zoonotic transfer (Denis et al. 2009; Van den Broek et al. 2009).
While the MRSA strains in North America (Smith et al. 2009) and Singapore (Sergio et al. 2007) are well documented, information on their counterparts in sub-Saharan Africa especially in the Eastern Cape Province of South Africa is very scarce; ditto for information on the antibiotic resistance properties of commensal bacteria of animal origin. In this study, the prevalence of staphylococci of farm animal origin in the Nkonkobe Municipality, Eastern Cape Province, South Africa as well as their phenotypic and genotypic resistance properties were investigated.
Materials and methods
Study location
The study location is the Nkonkobe Municipality, a highly populated domain in the Eastern Cape Province, South Africa, with a population of about 128,658 on a 3,724 km2 of land. The Staphylococcus spp. of farm animal origin were isolated from nasal, mouth wash, and ear swabs of pigs (including the piglets), cattle, goats, and chickens from various animal farms within the municipality. Some samples of the animal dung were also analyzed to see whether the isolates were shed to the environment in the animal dung.
Isolation and preliminary identification of Staphylococcus species
Preliminary isolation of Staphylococcus spp. of farm animal origin was initiated by inoculating the swabs directly in nutrient broth (CM0001B, Oxoid, England) and incubated at 37 °C for 24 h (Cheesebrough 2006). Thereafter, a loopful of inoculum was transferred from each turbid tube to mannitol salt agar (Oxoid, England) and incubated also for 24 h at 37 °C (Cheesebrough 2006). At the end of the incubation period, distinct colonies were purified and subjected to gram-staining, catalase test, coagulase test, and lysostaphin susceptibility test. The Staphylococci were preliminarily speciated using the analytical profile index (API) Staph (BioMerieux). The results were read following the incubation of inoculated strips between 18 and 24 h.
Genus- and specie-specific identification of Staphylococcus species
All isolates identified earlier using the API Staph kit were confirmed by genus-specific and species-specific polymerase chain reactions (PCR) using the primers listed in Table 1. The PCR conditions employed for the genus level identification include 3 min at 96 °C, 40 cycles of 30 s at 95 °C, 60 s at 55 °C, 30 s at 72 °C, and a final extension of 3 min at 72 °C (Martineau et al. 2001). S. aureus ATCC 25923 was used as the positive control, while nuclease-free water was used as the negative control. The amplicons were electrophoresed on 2.5 % agarose gel stained with ethidium bromide and visualized under UV light. Species-specific identification was done by multiplex PCR targeting Staphylococcus saprophyticus, Staphylococcus epidermidis, S. xylosus, and S. aureus (at the amplicon size in Table 1), and the PCR conditions include 15 min at 4 °C, 3 min at 94 °C, then 40 cycles of 1 s at 95 °C, 30 s at 55 °C, 30 s at 72 °C, and a final hold of 3 min at 72 °C (Corbière et al. 2004). The amplicons were electrophoresed in 1× TBE buffer on 2 % agarose gel stained with ethidium bromide and visualized under UV light. Other species were identified by simplex PCR with the conditions which include a denaturation step 10 min at 94 °C, followed by 35 cycles of 15 s at 94 °C, 30 s at the appropriate annealing temperature of 59, 50, and 60 °C for S. capitis, S. haemolyticus, and Staphylococcus warneri, respectively, and 30 s at 72 °C were employed (Iwase et al. 2007).
Phenotypic antibiotic susceptibility profile
The standard agar disk diffusion technique was employed to determine the antibiotic susceptibility pattern of the isolates, and this was performed in accordance with standards described by Cheesebrough (2006) and Clinical and Laboratory Standards Institute (CLSI) (2008). The antibiotics used include penicillin G (11 U), ampicillin (10 μg), oxytetracycline (10 μg), minocycline (10 μg), streptomycin (10 μg), cotrimoxazole (25 μg), cefotaxime (10 μg), erythromycin (10 μg), gentamicin (10 μg), clindamycin (2 μg), ceftriaxone (30 μg), oxacillin (5 μg), ceftriaxone (30 μg), ceftazidime (30 μg), vancomycin (5 μg), cephalothin (25 μg), imipenem (10 μg), meropenem (10 μg), ofloxacin (5 μg), levofloxacin (5 μg), and ciprofloxacin (5 μg). S. aureus ATCC 25923 was used as quality control. The results were interpreted according to the CLSI standards (NCCLS 1999; CLSI 2008).
Multiple antibiotic resistance index
The multiple antibiotic resistance index (MARI) was calculated as the ratio of the number of the antibiotics to which resistance occurred by the isolates (a) to the total number of antibiotics to which the isolates were tested (b) (Krumperman 1983).
All the antibiotics used were also considered for MARI determinations as organisms' exhibit unequal profile to antibiotics within the same class.
PCR detection of antibiotic resistance genes
Table 2 summarized the list of primers used for the PCR detection of ermA, ermB, ermC, msrA, and mphC genes following the protocol of Sauer et al. (2008) with slight modifications. The genomic DNA was extracted using PrepMan Ultra DNA extraction kit (Applied Biosystem). PCR cycles involve an initial denaturation step of 94 °C for 5 min followed by 30 amplification cycles including denaturation at 94 °C for 60 s, annealing at 51 °C for ermA, ermB, ermC, or 55 °C for msrA, mphC for 60 s, and extension at 72 °C for 60 s. A final extension at 72 °C for 5 min in 1 cycle then ended the PCR. For mecA, vanA, and vanB genes, PCR conditions include an initial denaturation step at 94 °C for 5 min, which will be followed by 10 cycles of amplification (denaturation at 94 °C for 30 s, annealing at 64 °C for 30 s, and extension at 72 °C for 45 s) and another 25 cycles of amplification (denaturation at 94 °C for 30 s, annealing at 50 °C for 45 s, and extension at 72 °C for 2 min), ending with a final extension step at 72 °C for 10 min.
Statistical analysis
The observed variables were converted into easily interpretable data. The significance of these data was determined using chi-square. A p value ≤0.05 was regarded as being statistically significant, while p values ≥0.05 was interpreted as being statistically nonsignificant (Dahiru 2008)
Results
Following phenotypic and molecular identification, 120 staphylococcal isolates of farm animal origin were recovered. The analytic profile index results showed well over 95 % agreement for genus-based identification, when compared with PCR-based genus-specific identification. Species-specific PCRs revealed the identity of the following staphylococcal species: S. xylosus (15 %), S. aureus (23.3 %), S. haemolyticus (35 %), S. capitis (15 %), and other Staphylococcus spp. (11.7 %) (Table 3).
For clarity, Table 3 specifically showed the recovery of Staphylococcus spp. with respect to their animal source(s). Sixty-four (55.6 %) of the isolates were recovered from pigs, 18 (15.7 %) from goats, and 24 (20.9 %) from cattle. These percentages refer to the quantity per total Staphylococcus spp. isolated. Twenty-eight (23.3 %) of the Staphylococcus spp. were coagulase-positive, while the remaining were coagulase-negative.
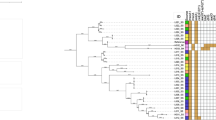
The results of antimicrobial susceptibility testing are shown in Table 4. Resistances of 40.2 % to ceftazidime, 75 % to penicillin G, 83.3 % to tetracycline, 100 % to nalidixic acid, and sulfamethoxazole were observed. Also, 38 % of the Staphylococcus spp. were resistant to oxacillin, while 12 % showed phenotypic resistance to vancomycin. The presence of the mecA genes was observed among the methicillin-resistant Staphylococcus spp. No vancomycin resistance genes (vanA and vanB) were detected in these organisms. Also, 12 % of the bacteria were resistant to erythromycin, while 40.2 % were resistant to ceftazidime. Over 68.4 % of the isolates were classified as multiresistant according to the MARI >2 (Fig. 1).
Discussion
The ability of staphylococci to survive in unfavorable osmotic conditions, pressure, and slightly elevated temperature supports their survival of staphylococci on animals (LeLoir et al. 2003). We were particularly interested in those species that can affect humans. In total, 30 % of the Staphylococci from pigs were S. haemolyticus and 23.3 % S. aureus. In our study, S. aureus has been only detected among the porcine isolates, which is in accordance with De Neeling et al. (2007). S. haemolyticus and some unidentified staphylococcal isolates were also observed from pig sources. S. capitis were isolated from goats only. S. capitis is a known flora of human scalp and skin, though it is also a frequently observed etiology of endocarditis (Van Der Zwet et al. 2002; Iwase et al. 2007; D'mello et al. 2008). About 5 to15% of the total staphylococcal isolates from the animal sources were S. haemolyticus and S. xylosus, respectively, and they were isolated from cattle. The total occurrences of S. haemolyticus observed in this study agrees with earlier reports on animal and animal products as reservoir of bacterial pathogens (Bagcigil et al. 2007; Schlegelova et al. 2008). Since there are at least 40 recognized Staphylococcus spp. (Trulzsch et al. 2002; Bannerman 2003), assaying for all of them may not be feasible in this kind of study (Deurenberg and Stobberingh 2008). Hence, 11.7 % of the Staphylococcus spp. identified to genus level and recovered from pigs and chicken could not be speciated into any of the target species listed in Table 3. The recovery of more CoNS (76.7 %) corroborates previous reports (Gortel et al. 1999; Kania et al. 2004). The elaboration of staphylocoagulase, as observed in this study, has been described as a very important factor in determining the inherent pathogenicity of bacteria also in commensal flora (Fairbrother 2005).
The observed resistance patterns to some of the conventional antibiotics which are usually frequently prescribed in the study area are alarming because of the high resistance rates among nonclinical isolates. This further reaffirms the critical role of commensals in public health. The observed high level (75–100 %) of resistance to sulfamethoxazole, nalidixic acid, tetracycline, and improved penicillin G, all of which are broad-spectrum antibiotics, might be due to consumption of antimicrobials (Moulin 2001) as growth promoters (Perrier-Gros-Claude et al. 1998), as extensively practiced in the study area. Of the 38 % resistance to methicillin/oxacillin observed, 12.6 % was S. aureus from pig. Though oxacillin is more stable than methicillin during storage, laboratory diagnosis of methicillin resistance depends on the testing of oxacillin, and methicillin/oxacillin-resistant staphylococcal isolates are supposed to be reported as being resistant to β-lactam antibiotics (CLSI 2008). Vancomycin used to be the last antibiotic for treating infections caused by such resistant isolates (Fitzgerald et al. 2001; Boucher et al. 2010). In fact, it was a drug in the last line of defense (Bhalakia and Morris 2005). In this study, 12 % of the staphylococcal isolates were vancomycin-resistant and were recovered from all animal species investigated in this study. Unfortunately, there is usually a close link between the resistance to vancomycin and to other extended spectrum beta-lactam drugs like meropenem and imipenem, with a tendency to worsen the difficulty in the choice of therapeutic options (Chang et al. 2003; Boucher et al. 2010). Tenover and Goering (2009) also reported the presence of community-based MRSA, just as Bhalakia and Morris (2005) also reported the presence of plasmid-mediated vancomycin resistance in fomite. The MAR index of >0.2 observed in this study suggests that the isolates emerged from high-risk sources (Suresh et al. 2000).
The observed phenotypic methicillin/oxacillin resistance in this study was confirmed by the detection of the mecA genes. The mecA gene confers resistance against β-lactamase-stable penicillin and virtually all other β-lactam antibiotics (Pinho et al. 2001; Weese et al. 2005). The mecA gene codes for an alternative penicillin binding protein PBP2a which has low affinity for β-lactam antibiotics (Pinho et al. 2001). The observed absence of mecA gene among few methicillin-resistant Staphylococcus spp. in this study corroborates the observation of Montanari et al. (1990) that the presence of underlining factors for methicillin resistance apart from mecA genes. The presence of this gene in the commensal organisms might render them difficult to control, given the opportunity to zoonotically infect immunocompromised individuals. This gene is usually housed in a large mobile genetic element known as staphylococcal cassette chromosome mec (SCCmec) (Weese et al. 2005). Meanwhile, at least 11 different SCCmec types have been described, and some of them are found in humans (Kriegeskorte et al. 2012). This has been described as hospital-associated and/or community-associated MRSA (Otter and French 2010). We, therefore, proposed that the observed MRSA in the pig and methicillin-resistant S. haemolyticus could also have been transferred from human sources to the animals, as they are also possible colonizers of human hosts. Transfer to these animals from man through an intermediate animal is also a possibility. However, the presence of the mecA gene gives a genetic basis for resistance of the Staphylococcus spp. to methicillin and other lower beta-lactam antibiotics (Duquette and Nuttall 2004), even if phenotypic susceptibility to beta-lactam is seemingly observed.
Lower resistance (12 %) of the Staphylococcus spp. to erythromycin was observed than to the third generation cephalosporin, ceftazidime (40.2 %). This confirms the earlier report by Essack (2001) that some lower class β-lactam antibiotics showed better activities to gram-positive bacteria than third generation cephalosporin. The detection of mphC gene justifies the phenotypic resistance to erythromycin and serves as representative of such genes among commensals in the study area. Meanwhile, the wide range of multiple antibiotic resistance indices showed a divergence between the static-use (in situation where a fixed antibiotic regimen is used) and the adaptive-use (in using varieties or wide range of antibiotics due to observed low performance of the earlier one(s)), which may imply consistent use of various antibiotics in these farms on the animal, to achieve a non-chemotherapeutic advantage (Laxminarayan and Klugman 2011). This implies that the organisms might have developed resistance over a period of exposure without medical prescription. An observation of MAR index >0.2 means that the isolate source is high risk, which is a source where antibiotics are in constant abuse, and the act is bringing about high selective pressure (Suresh et al. 2000).
On the animal impact on the environment, the presence of these isolates in the animal dung showed that the mecA-positive isolates are released to the environment by these organisms. This corroborates the observation of Alexander et al. (2011).
Therefore, this exposure of the animal bacterial flora to antibiotics appears to be an encouraging emergence of resistance across a wide range of antibiotics (Barbosa and Levy 2000). It is, therefore, important to control the misuse or any other non-therapeutic use of antibiotics. This study is, therefore, important to the agriculture sector in South Africa as animal health is involved. It is also important to the health sector due to the possibility of zoonotic infections, if care is not taken, even if the organism in the animals has its original source from previous human contact. Piggery workers should be diligently hygienic as the animal is a consistent source of MRSA. Regular PCR-based assessment of MRSA prevalence in various aspects of natural life and hospitals is hereby advocated to bring about appropriate control strategies and to reduce the present scourge of MRSA in multidrug resistant outbreaks.
References
Alexander TA, Yanke JL, Reuter T, Topp E, Read RR, Selinger BL, McAllister TA (2011) Longitudinal characterization of antimicrobial resistance genes in feces shed from cattle fed different subtherapeutic antibiotics. BMC Microbiol 11:19. doi:10.1186/1471-2180-11-19
Bagcigil AF, Moodley A, Jensen VE, Guardabassi L (2007) Occurrence, species distribution, antimicrobial resistance, and clonality of methicillin- and erythromycin-resistant staphylococci in the nasal cavity of domestic animals. Vet Microbiol 121:307–315
Bannerman TL (2003) Staphylococcus, Micrococcus, and other catalase-positive cocci that grow aerobically. In: Murray PR, Baron EJ, Jorgensen JH, Pfaller MA, Yolken RH (eds) Manual of Clinical Microbiology. American Society Microbiology, Washington, pp 384–404
Barbosa TM, Levy SB (2000) The impact of antibiotic use on resistance development and persistence. Drug Res Updates 3:303–311
Bhalakia N, Morris D (2005) Isolation and Plasmid Analysis of Vancomycin- Resistant Staphylococcus aureus. J Young Invest. 13 (4), retrieved 11 December 2010 from http://www.jyi.org/research/re.php?id=573
Boucher H, Miller LG, Razonable RR (2010) Serious infections caused by methicillin-resistant Staphylococcus aureus. Clin Infect Dis 51(Suppl 2):S183–S197
Chang S, Sievert DM, Hageman JC (2003) Infection with vancomycin resistant Staphylococcus aureus containing the vanA resistance gene. N Engl J Med 348:1342–1347
Cheesebrough M (2006) District Laboratory Practice in Tropical Countries. Part 2, 2nd edition, Cambridge University Press pp. 132–143.
Clinical and Laboratory Standards Institute, CLSI (2008) Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals—Third Edition: Approved Standard M31-A3. CLSI, Wayne, PA, USA
Cohen ML (1992) Epidemiology of drug resistance: implications for a post antimicrobial era. Science 257:1050–1055
Corbière MS, Talon R, Leroy S (2004) Development of a multiplex PCR for the identification of Staphylococcus genus and four staphylococcal species isolated from food. J Appl Microbiol 97:1087–1094
Cuny C, Friedrich A, Kozytska S, Layer F, Nubel U et al (2010) Emergence of methicillin-resistant Staphylococcus aureus (MRSA) in different animal species. Int J Med Microbiol 300(2–3):109–117
Dahiru T (2008) P value, a true test of statistical significance? A cautionary note. Ann. Ibad. Postgrad. Medicine 6(1):21–26
De Neeling AJ, van den Broek MJM, Spalburg EC, van Santen-Verheuvel MG, Dam-Deisz WDC et al (2007) High prevalence of methicillin-resistant Staphylococcus aureus in pigs. Vet Microbiol 122:366–372
Denis O, Suetens C, Hallin M, Catry B, Ramboer I et al (2009) Methicillin-resistant Staphylococcus aureus ST398 in swine farm personnel, Belgium. Emerg Infect Dis 15(7):1098–1101
Deurenberg RH, Stobberingh EE (2008) “The evolution of Staphylococcus aureus” available at <http://www.ncbi.nlm.nih.gov/pubmed/18718557>
D'mello D, Daley AJ, Rahman MS, Garland S, Pearce C et al (2008) Vancomycin heteroresistance in bloodstream isolates of Staphylococcus capitis. J Clin Microbiol 46(9):3124–3126
Duquette RA, Nuttall TJ (2004) Methicillin-resistant Staphylococcus aureus in dogs and cats: an emerging problem? J Small Anim Pract 45(12):591–597
Dutka-Malen S, Evers S, Courvalin P (1995) Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol 33:24–27
Essack SY (2001) The development of β-lactam antibiotics in response to the evolution of β-lactamases. Pharm Res 18(10):1391–1399
Fairbrother RW (2005) Coagulase production as a criterion for the classification of the staphylococci. J Pathol Bacteriol 50(1):83–88
Fitzgerald JR, Sturdevant DE, Mackie SM, Gill SR, Musser JM (2001) Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc Natl Acad Sci U S A 98(15):8821–8826
Geha DJ, Uhl JR, Gustaferro CA, Persing DH (1994) Multiplex PCR for identification of methicillin-resistant staphylococci in the clinical laboratory. J Clin Microbiol 32:1768–1772
Gortel K, Campbell KL, Kakoma I, Whittem T, Schaeffer DJ et al (1999) Methicillin resistance among staphylococci isolated from dogs. Am J Vet Res 60:1526–1530
Grundmann H, Aires-de-Sousa M, Boyce J, Tiemersma E (2006) Emergence and resurgence of methicillin-resistant Staphylococcus aureus as a public-health threat. Lancet 368(9538):874–885
Hammerum AM, Lester CH, Heuer OE (2010) Antimicrobial-resistant enterococci in animals and meat: a human health hazard? Foodborne Pathog Dis 7(10):1137–46
Harada K, Asai T (2010) Role of antimicrobial selective pressure and secondary factors on antimicrobial resistance prevalence in Escherichia coli from food-producing animals in Japan. J Biomed Biotechnol. doi:10.1155/2010/180682
Iwase T, Seki K, Shinji H, Mizunoe Y, Masuda S (2007) Development of a real- time PCR assay for the detection and identification of Staphylococcus capitis, Staphylococcus haemolyticus, and Staphylococcus warneri". J Med Microbiol 56(Pt 10):1346–1349
Jensen LB, Frimodt-Moller N, Aarestrup FM (1999) Presence of erm gene classes in gram-positive bacteria of animal and human origin in Denmark. FEMS Microbiol Lett 170:151–158
Kania SA, Williamson NL, Frank LA, Wilkes RP, Jones RD et al (2004) Methicillin resistance of staphylococci isolated from the skin of dogs with pyoderma. Am J Vet Res 65:1265–1268
Kaur R, Pathania R (2010) Drug Resistance in Food Animals - A Public Health Concern. Onl. Vet J. 5(1), Article 48. Available at: <http://www.vetscan.co.in/v5n1/drug_resistance_in_food_animals_a_public_health _concern.htm>
Kitara LD, Anywar AD, Acullu D, Odongo-Aginya E, Aloyo J et al (2011) Antibiotic susceptibility of Staphylococcus aureus in suppurative lesions in Lacor Hospital. Uganda Afr Health Sci 11(S1):S34–S39
Kloos WE, Bannerman TL (1994) Update on clinical significance of coagulase-negative staphylococci. Clin Microbiol Rev 7(1):117–140
Komolafe AO, Adegoke AA (2008) Incidence of bacterial septicemia in Ile-Ife Metropolis, Malaysian J. Microbiology 4(2):51–61
Kriegeskorte A, Ballhausen B, Idelevich AE, Kock R, Friedrich AW, Karch H, Peters G, Becker K (2012) Human MRSA isolates with novel genetic homolog, Germany. Emerg Infec Dis 18(6):1016–1018
Krumperman PH (1983) Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol 46(1):165–170
Laxminarayan R, Klugman KP (2011) Communicating trends in resistance using a drug resistance index. BMJ Open;1: e000135 doi:10.1136/bmjopen-2011-000135.
LeLoir Y, Baron F, Gautier M (2003) Staphylococcus aureus and food poisoning. Gen Mol Res 2(1):63–76
Levy SB (1986) Ecology of antibiotic resistance determinants. In: Press CSHL (ed) Antibiotic resistance genes: ecology, transfer and expression. Cold Spring Harbor Press, New York, pp 17–30
Lüthje P, Schwarz S (2006) Antimicrobial resistance of coagulase-negative Staphylococci from bovine subclinical mastitis with particular reference to macrolide-lincosamide resistance phenotypes and genotypes. J Antimicrob Chemother 57:966–969
Martineau F, Picard FJ, Ke D, Paradis S, Roy PH et al (2001) Development of a PCR assay for identification of staphylococci at genus and species levels. J Clinical Microbiol 39:2541–2547
Montanari MP, Tonin E, Biavasco F, Varaldo PE (1990) Further characterization of borderline methicillin-resistant Staphylococcus aureus and analysis of penicillin-binding proteins. Antimicrob Agents Chemother 34:911–913
Morot-Bizot SC, Talon R, Leroy S (2004) Development of a multiplex PCR for the identification of Staphylococcus genus and four staphylococcal species isolated from food. J Appl Microbiol 97:1087–1094
Moulin G (2001) Surveillance of antimicrobial consumption : activities in France (Agence Nationale du Médicament Vétérinaire). In: 2nd International Conference of the Office International des Epizoosties, 2001; Paris; 2001.
NCCLS (1999) Performance Standards for Antimicrobial Disk and Dilution Susceptibility, Tests for Bacteria Isolated from Animals; Approved Standard. NCCLS Document M31-A .
Ogbodo SO, Okeke AC, Ugwuoru CDC, Chukwurah EF (2011) Possible Alternatives to Reduce Antibiotic Resistance. Life Sc. Med Res available at: <http://astonjournals.com/lsmr> [Accessed 12 December 2011]
Otter JA, French GL (2010) Molecular epidemiology of community-associated methicillin-resistant Staphylococcus aureus in Europe. Lancet Infect Dis 10(4):227–239
Perrier-Gros-Claude J, Courrier P, Bréard J, Vignot J, Masseront T et al (1998) Entérocoques résistants aux glycopeptides dans les viandes. Bull Epidemiol Hebdomadaire 50–51
Pinho MG, de Lencastre H, Tomasz A (2001) An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc Natl Acad Sci U S A 98(19):10886–10891
Ramos-Trujillo E, Perez-Roth E, Mendez-Alvarez S, Claverie-Martin F (2003) Multiplex PCR for simultaneous detection of enterococcal genes vanA and vanB and staphylococcal genes mecA, ileS-2, and femB. Int Microbiol 6:113–115
Saha B, Singh AK, Abhrajyoti G, Bal M (2007) Identification and characterization of a vancomycin resistant Staphylococcus aureus isolated from Kolkata (South Asia). J Med Microbiol 57:72–79
Saleha A, Zunita Z (2010) Methicillin-resistant Staphylococcus aureus (MRSA): An emerging veterinary and zoonotic pathogen of public health concern and some studies in Malaysia. J Anim Vet Adv 9(7):1094–1098
Sauer P, Sila J, Stosova T, Vecerova R, Hejnar P et al (2008) Prevalence of genes encoding extracellular factors among methicillin-resistant Staphylococcus aureus isolates from the University Hospital, Olomouc Czech Republic. J Med Microbiol 57:403–410
Schlegelova J, Vlkova H, Babak V, Holasova M, Jaglic Z et al (2008) Resistance to erythromycin of Staphylococcus spp. isolates from the food chain. Vet Med 53(6):307–314
Sergio DM, Koh TH, Hsu LY, Ogden BE, Goh AL et al (2007) Investigation of methicillin-resistant Staphylococcus aureus in pigs used for research. J Med Microbiol 56(Pt 8):1107–1109
Shittu A, Lin J, Morrison D, Kolawole D (2004) Isolation and molecular characterization of multiresistant Staphylococcus sciuri and Staphylococcus haemolyticus associated with skin and soft-tissue infections. J Med Microbiol 53(1):51–55
Smith TC, Male MJ, Harper AL, Kroeger JS, Tinkler GP et al (2009) Methicillin- resistant Staphylococcus aureus (MRSA) strain ST398 is present in Midwestern US swine and swine workers. PLoS One 4(1):e4258
Summers AO (2002) Generally overlooked fundamentals of bacterial genetics and ecology. Clin Infect Dis 34(Suppl 3):S85–S92
Suresh T, Srinivasan D, Hatha AAM, Lakshmanaperumalsamy P (2000) The incidence, antibiotics resistance, and survival of Salmonella and Escherichia coli isolated from broiler chicken retail outlets. Microb Environ 15:173–181
Tenover FC, Goering RV (2009) Methicillin-resistant Staphylococcus aureus strain USA300: origin and epidemiology. J Antimicrob Chemother 64(3):441–446
Thorberg B (2008) Coagulase-Negative Staphylococci in Bovine Sub-Clinical Mastitis. Licentiate Thesis Department of Biomedical Sciences and Veterinary Public Health Swedish University of Agricultural Sciences Report no. 2
Trulzsch K, Rinder H, Treek J, Bader L, Wilhelm U et al (2002) "Staphylococcus pettenkoferi", a novel Staphylococcal species isolated from clinical specimens. Diag Microbiol Infect Dis 43:175–182
Van den Broek IV, Van Cleef BA, Haenen A, Broens EM, Van der Wolf PJ et al (2009) Methicillin-resistant Staphylococcus aureus in people living and working in pig farms. Epidemiol Infect 137(5):700–708
Van Der Zwet WC, Debets-Ossenkopp YJ, Reinders E, Kapl M, Savelkoul PHM et al (2002) Nosocomial spread of a Staphylococcus capitis strain with hetero-resistance to vancomycin in a neonatal intensive care unit. J Clin Microbiol 40(7):2520–2525
Van Loo I, Huijsdens X, Tiemersma E, de Neeling A, van de Sande-Bruinsma N et al (2007) Emergence of methicillin-resistant Staphylococcus aureus of animal origin. Emerg Infect Dis 13(12):1834–1839
Walther B, Wieler LH, Friedrich AW, Hanssen AM, Kohn B, Brunnberg L, Lubke-Becker A (2008) Methicillin-resistant Staphylococcus aureus (MRSA) isolated from small and exotic animals at a university hospital during routing microbiological examinations. Vet Microbiol 127(1–2):171–178
Weese JS, Archambault M, Willey BM, Hearn P, Kreiswirth BN et al (2005) Methicillin-resistant Staphylococcus aureus in horses and horse personnel, 2000–2002. Emerg Infect Dis 11(3):430–435
Werckenthin C, Schwarz S (2000) Molecular analysis of the translational attenuator of a constitutively expressed ermA gene from Staphylococcus intermedius. J Antimicrob Chemother 46:785–788
WHO (2011) Removing obstacles to healthy development. http://www.who.int/infectious-disease-report/pages/textonly.html#Anchor1. [Accessed 10 December 2011]
Wondrack L, Massa M, Yang V, Sutcliffe J (1996) Clinical strain of Staphylococcus aureus inactivates and causes efflux of macrolides. Antimicrob Agents Chemother 40:992–998
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Adegoke, A.A., Okoh, A.I. Species diversity and antibiotic resistance properties of Staphylococcus of farm animal origin in Nkonkobe Municipality, South Africa. Folia Microbiol 59, 133–140 (2014). https://doi.org/10.1007/s12223-013-0275-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-013-0275-1