Abstract
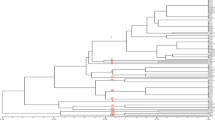
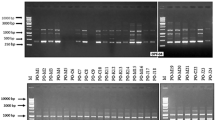
Genetic diversity of 89 isolates of Rhizoctonia solani isolated from different pulse crops representing 21 states from 16 agro-ecological regions of India, 49 morphological, and 7 anastomosis groups (AGs) was analyzed using 12 universal rice primers (URPs), 22 random amplified polymorphic DNA (RAPD), and 23 inter-simple sequence repeats (ISSR) markers. Both URPs and RAPD markers provided 100 % polymorphism with the bands ranging from 0.1 to 5 kb in size, whereas ISSR markers gave 99.7 % polymorphism with the bands sizes ranging from 0.1 to 3 kb. The marker URP 38F followed by URP13R, URP25F, and URP30F, RAPD marker R1 followed by OPM6, A3 and OPA12 and ISSR3 followed by ISSR1, ISSR4, and ISSR20 produced the highest number of amplicons. R. solani isolates showed a high level of genetic diversity. Unweighted pair group method with an arithmetic average (UPGMA) analysis grouped the isolates into 7 major clusters at 35 % genetic similarity using the three sets of markers evaluated. In spite of using three different types of markers, about 95 % isolates shared common grouping patterns. The majority of the isolates representing various AGs were grouped together into different sub-clusters using all three types of markers. Molecular groups of the isolates did not correspond to agro-ecological regions or states and crops of the origin. An attempt was made for the first time in the present study to determine the genetic diversity of R. solani populations isolated from different pulse crops representing various AGs and agro-ecological regions.







Similar content being viewed by others
References
Aggarwal R, Aradhika T, Yadav A (2010) Pathogenic and genetic variability in Tilletia indica monosporidial culture lines using universal rice primer-PCR. Eur J Plant Pathol 128:333–342
Agricultural Statistics at a Glance (2010) Directorate of Economics and Statistics, Department of Agriculture, Ministry of Agriculture, Government of India
Baitamar RM, Salari M, Zafari D (2010) Anastomosis groups of Rhizoctonia solani from potato and survey of somatic compatible groups by morphological characters and RAPD marker. Iran J Plant Pathol 45:87–89
Carling DE (1996) Grouping in Rhizoctonia solani by hyphal anastomosis. In: Sneh B, Jabaji-Hare S, Neate S, Dijst G (eds) Rhizoctonia Species: taxonomy, molecular biology, ecology, pathology and disease control. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 37–47
Cobb BD, Clarkson JM (1994) A simple procedure for optimizing the polymerase chain reaction (PCR) using modified Tuguchi methods. Nucl Acid Res 22:3801–3805
Cubeta MA, Echandi E et al (1991) Characterization of anastomosis groups of binucleate Rhizoctonia species using restriction analysis of an amplified ribosomal RNA gene. Phytopathology 81:272–280
Dubey SC (2003) Integrated management of web blight of urd/mung bean by bio-seed treatment. Indian Phytopath 56:34–38
Dubey SC, Dwivedi RP (2000) Diseases of leguminous crops caused by Rhizoctonia solani and their management. In: Narain Udit, Kumar K, Srivastava M (eds) Advances in plant disease management. Advance Publishing Concept, New Delhi, pp 61–76
Dubey SC, Singh SR (2008) Virulence analysis and oligonucleotide fingerprinting to detect diversity among Indian isolates of Fusarium oxysporum f. sp. ciceris causing chickpea wilt. Mycopathologia 165:389–406
Duncan S, Barton JE, O’Brien PA (1993) Analysis of variation in isolates of Rhizoctonia solani by random amplified polymorphic DNA assay. Mycol Res 97:1075–1082
Fenille RC, de Souza NL, Kuramae EE (2002) Characterization of Rhizoctonia solani associated with soybean in Brazil. Eur J Plant Pathol 108:783–792
Hwang SF, Gossen BD et al (2003) Etiology, impact and control of Rhizoctonia seedling blight and root rot of chickpea on the Canadian prairies. Can J Plant Sci 83:959–967
Justesen AF, Yohalem D et al (2003) Genetic diversity in potato field populations of Thanatephorus cucumeris AG-3, revealed by ITS polymorphism and RAPD markers. Mycol Res 107:1323–1331
Kang HW, Park DS et al (2002) Fingerprinting of diverse genomes using PCR with universal rice primers generated from repetitive sequence of Korean weedy rice. Mol Cell 13:281–287
Khodayari M, Safaie N, Shamsbakhsh M (2009) Genetic diversity of Iranian AG1-IA isolates of Rhizoctonia solani, the cause of rice sheath blight, using morphological and molecular markers. J Phytopathol 157:708–714
Kronland WC, Stanghellini ME (1988) Clean slide technique for the observation of anastomosis and nuclear condition of Rhizoctonia solani. Phytopathology 78:820–822
Kuninaga S, Natsuaki T et al (1997) Sequence variation of the rDNA ITS regions within and between anastomosis groups in Rhizoctonia solani. Curr Genet 32:237–243
Lubeck M (2004) Molecular characterization of Rhizoctonia solani. Appl Mycol Biotech 4:205–224
Murray MG, Thompson WE (1980) Rapid isolation of high molecular weight plant DNA. Nucl Acid Res 8:4321–4325
Ogoshi A (1987) Ecology and pathogenicity of anastomosis and intraspecific groups of Rhizoctonia solani Kühn. Ann Rev Phytopathol 25:125–143
Parmeter JR Jr, Whiteny HS (1970) Taxonomy and nomenclature of the imperfect state. In: Parmeter JR Jr (ed) Rhizoctonia solani: biology and pathology. California Press, Berkeley, pp 20–31
Parmeter JR Jr, Sherwood RT, Platt WD (1969) Anastomosis grouping among isolates of Thanatephorus cucumeris. Phytopathology 59:1270–1278
Rohlf JF (1998) NTSYS numerical taxonomy and multivariate analysis system version 2.02
Saksena HK, Vaartaja O (1961) Taxonomy, morphology and pathogenicity of Rhizoctonia species from forest nurseries. Can J Bot 39:627–647
Sharma M, Gupta SK, Sharma TR (2005) Characterization of variability in Rhizoctonia solani by using morphological and molecular markers. J Phytopathol 153:449–456
Sneh B, Burpee L, Ogoshi A (1991) Identification of Rhizoctonia species. APS Press, St. Paul, MN., USA, p 133
Sneh B, Jabaji-Hare S et al (1996) Rhizoctonia species: taxonomy, molecular biology, ecology, pathology, and disease control. Kluwer Academic Publishers, Dordrecht, the Netherlands
Williams JGK, Kubelik AR et al (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucl Acid Res 18:6531–6535
Yang J, Kharbanda PD et al (1996) Characterization, virulence, and genetic variation of Rhizoctonia solani AG-9 in Alberta. Plant Dis 80:513–518
Zhou Q, Chang K et al (2009) Pathogenicity and genetic diversity of Rhizoctonia solani isolates from lupin and other crops in Alberta, Canada. Can J Plant Pathol 31:340–347
Acknowledgments
Authors are thankful to Indian Council of Agricultural Research, New Delhi for financial support and Dr. H. Nirenberg, Julius Kühn- Institute (BBA), Germany and Dr. Mitsuro Hyakumachi, Gifu University, Japan for providing international tester isolates of various AGs.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dubey, S.C., Tripathi, A. & Upadhyay, B.K. Molecular diversity analysis of Rhizoctonia solani isolates infecting various pulse crops in different agro-ecological regions of India. Folia Microbiol 57, 513–524 (2012). https://doi.org/10.1007/s12223-012-0165-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-012-0165-y