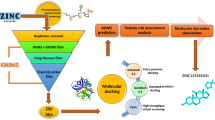
Abstract
A further refinement to our previously published “target-bound pharmacophore modeling approach” is reported herein. Instead of relying on “random” pharmacophores—which is the most commonly used approach in literature—phrmacophoric “hot spots” were selected based on highly contributing amino acid residues to inhibitor binding. Highly contributing amino acid residues were identified based on energy footprints obtained from molecular dynamics and thermodynamic calculations. To our knowledge, this is the first attempt in the literature to use this approach. Previously, docking energy was used to create energy-based pharmacophores, however, docking scores, in many cases, are artifacts and questionable. Hence, we introduce the concept of per-residue energy decomposition footprints obtained from molecular dynamics ensembles in order to create more reliable pharmacophore modeling which can be implemented in drug discovery workflows. The purpose of this work is not to compare the validity of this approach against previously reported ones, but rather to introduce a more rational approach to the scientific domain. Further computational and experimental validations would be advised. Herein, we applied the developed protocol in order to identify novel BACE1 inhibitors and results were validated against experimentally determined inhibitors with known activity against BACE1. This protocol was found promising to identify hits with higher binding affinities when compared against experimentally determined inhibitors. Other post-dynamic binding analysis for the identified hits was also presented in this report as a roadmap towards the design of potential BACE1 inhibitors as anti-Alzheimer Agents.









Similar content being viewed by others
References
Amaravadhi, H., K. Baek, and H. S. Yoon. Revisiting de novo drug design: receptor based pharmacophore screening. Curr. Top. Med. Chem. 14:1890–1898, 2014.
Anand, R., K. D. Gill, and A. A. Mahdi. Therapeutics of Alzheimer’s disease: past, present and future. Neuropharmacology 76:27–50, 2014.
Awale, M., X. Jin, and J. L. Reymond. Stereoselective virtual screening of the ZINC database using atom pair 3D-fingerprints. J. Cheminform. 7:3, 2015.
Awale, M., and J. L. Reymond. A multi-fingerprint browser for the ZINC database. Nucleic Acids Res. 42:W234–W239, 2014.
Batool, S., Z. A. Khan, W. Kamal, and M. A. Kamal. In silico screening for identification of novel anti-malarial inhibitors by molecular docking, pharmacophore modeling and virtual screening. Med. Chem. 11:687–700, 2015.
Bhakat, S., S. Chetty, A. J. Martin, and M. E. Soliman. Multi-drug resistance profile of PR20 HIV-1 protease is attributed to distorted conformational and drug binding landscape: molecular dynamics insights. J. Biomol. Struct. Dyn. 11:1–46, 2015.
Bhakat, S., A. J. M. Martin, and M. E. S. Soliman. An integrated molecular dynamics, principal component analysis and residue interaction network approach reveals the impact of M184V mutation on HIV reverse transcriptase resistance to lamivudine. Mol. Biosyst. 10(8):2215–2228, 2014. doi:10.1039/c4mb00253a
Bolognesi, M. L., R. Matera, A. Minarini, M. Rosini, and C. Melchiorre. Alzheimer’s disease: new approaches to drug discovery. Curr. Opin. Chem. Biol. 13:303–308, 2009.
Bowers, S., Y.-Z. Xu, S. Yuan, G. D. Probst, R. K. Hom, W. Chan, A. W. Konradi, H. L. Sham, Y. L. Zhu, P. Beroza, H. Pan, E. Brecht, N. Yao, J. Lougheed, D. Tam, Z. Ren, L. Ruslim, M. P. Bova, and D. R. Artis. Structure-based design of novel dihydroisoquinoline BACE-1 inhibitors that do not engage the catalytic aspartates. Bioorg. Med. Chem. Lett. 23:2181–2186, 2013.
Braga, R. C., and C. H. Andrade. Assessing the performance of 3D pharmacophore models in virtual screening: how good are they? Curr. Top. Med. Chem. 13:1127–1138, 2013.
B-Rao, C., A. Kulkarni-Almeida, K. V. Katkar, S. Khanna, U. Ghosh, A. Keche, P. Shah, A. Srivastava, V. Korde, K. V. S. Nemmani, N. J. Deshmukh, A. Dixit, M. K. Brahma, U. Bahirat, L. Doshi, R. Sharma, and H. Sivaramakrishnan. Identification of novel isocytosine derivatives as xanthine oxidase inhibitors from a set of virtual screening hits. Bioorg. Med. Chem. 20:2930–2939, 2012.
Butini, S., E. Gabellieri, M. Brindisi, A. Casagni, E. Guarino, P. B. Huleatt, N. Relitti, V. La Pietra, L. Marinelli, M. Giustiniano, E. Novellino, G. Campiani, and S. Gemma. Novel peptidomimetics as BACE-1 inhibitors: synthesis, molecular modeling, and biological studies. Bioorg. Med. Chem. Lett. 23:85–89, 2013.
Camps, P., X. Formosa, C. Galdeano, D. Munoz-Torrero, L. Ramirez, E. Gomez, N. Isambert, R. Lavilla, A. Badia, M. V. Clos, M. Bartolini, F. Mancini, V. Andrisano, M. P. Arce, M. I. Rodriguez-Franco, O. Huertas, T. Dafni, and F. J. Luque. Pyrano[3,2-c]quinoline-6-chlorotacrine hybrids as a novel family of acetylcholinesterase- and beta-amyloid-directed anti-Alzheimer compounds. J. Med. Chem. 52:5365–5379, 2009.
Chang, W. P., X. Huang, D. Downs, J. R. Cirrito, G. Koelsch, D. M. Holtzman, A. K. Ghosh, and J. Tang. Beta-secretase inhibitor GRL-8234 rescues age-related cognitive decline in APP transgenic mice. FASEB J. 25:775–784, 2011.
Chiriano, G., A. De Simone, F. Mancini, D. I. Perez, A. Cavalli, M. L. Bolognesi, G. Legname, A. Martinez, V. Andrisano, P. Carloni, and M. Roberti. A small chemical library of 2-aminoimidazole derivatives as BACE-1 inhibitors: structure-based design, synthesis, and biological evaluation. Eur. J. Med. Chem. 48:206–213, 2012.
Christopeit, T., G. Stenberg, T. Gossas, S. Nyström, V. Baraznenok, E. Lindström, and U. H. Danielson. A surface plasmon resonance-based biosensor with full-length BACE1 in a reconstituted membrane. Anal. Biochem. 414:14–22, 2011.
Csukly, G., E. Siraly, Z. Hidasi, P. Salacz, A. Szabo, and E. Csibri. Pharmacological and other options in preventing dementia: a literature review. Neuropsychopharmacol. Hung. 16:121–126, 2014.
Czech, C., and F. Grueninger. Animal models for Alzheimer’s disease—the industry perspective. Drug Discovery Today 10:e73–e78, 2013.
Dessolin, J. N-(3-(2-amino-6,6-difluoro-4,4a,5,6,7,7a-hexahydro-cyclopenta[e][1,3]oxazin-4-yl) -phenyl)-amides as BACE1 inhibitors: a patent evaluation of WO2013041499. Expert Opin. Ther. Pat. 24:239–242, 2014.
Dhanjal, J. K., S. Goyal, S. Sharma, R. Hamid, and A. Grover. Mechanistic insights into mode of action of potent natural antagonists of BACE-1 for checking Alzheimer’s plaque pathology. Biochem. Biophys. Res. Commun. 443:1054–1059, 2014.
Duan, X., M. Zhang, X. Zhang, F. Wang, and M. Lei. Molecular modeling and docking study on dopamine D2-like and serotonin 5-HT2A receptors. J. Mol. Graph. Model. 57:143–155, 2015.
Fernandez-Bachiller, M. I., A. Horatscheck, M. Lisurek, and J. Rademann. Alzheimer’s disease: identification and development of beta-secretase (BACE-1) binding fragments and inhibitors by dynamic ligation screening (DLS). ChemMedChem 8:1041–1056, 2013.
Ferreira, L. G., R. N. Dos Santos, G. Oliva, and A. D. Andricopulo. Molecular Docking and Structure-Based Drug Design Strategies. Molecules 20:13384–13421, 2015.
Ghosh, A. K., S. Gemma, and J. Tang. β-Secretase as a therapeutic target for Alzheimer’s disease. Neurotherapeutics 5:399–408, 2008.
Ghosh, A. K., N. Kumaragurubaran, L. Hong, S. Kulkarni, X. Xu, H. B. Miller, D. Srinivasa Reddy, V. Weerasena, R. Turner, W. Chang, G. Koelsch, and J. Tang. Potent memapsin 2 (β-secretase) inhibitors: design, synthesis, protein-ligand X-ray structure, and in vivo evaluation. Bioorg. Med. Chem. Lett. 18:1031–1036, 2008.
Halperin, I., B. Ma, H. Wolfson, and R. Nussinov. Principles of docking: an overview of search algorithms and a guide to scoring functions. Proteins 47:409–443, 2002.
Hanessian, S., Z. Shao, C. Betschart, J.-M. Rondeau, U. Neumann, and M. Tintelnot-Blomley. Structure-based design and synthesis of novel P2/P3 modified, non-peptidic β-secretase (BACE-1) inhibitors. Bioorg. Med. Chem. Lett. 20:1924–1927, 2010.
Hossain, T., A. Mukherjee, and A. Saha. Chemometric design to explore pharmacophore features of BACE inhibitors for controlling Alzheimer’s disease. Mol. Biosyst. 11:549–557, 2015.
Huang, D., Y. Liu, B. Shi, Y. Li, G. Wang, and G. Liang. Comprehensive 3D-QSAR and binding mode of BACE-1 inhibitors using R-group search and molecular docking. J. Mol. Graph. Model. 45:65–83, 2013.
Huey, R., G. M. Morris, A. J. Olson, and D. S. Goodsell. A semiempirical free energy force field with charge-based desolvation. J. Comput. Chem. 28:1145–1152, 2007.
Jain, A. N. Virtual screening in lead discovery and optimization. Curr. Opin. Drug Discov. Devel. 7:396–403, 2004.
John, S., S. Thangapandian, S. Sakkiah, and K. W. Lee. Potent BACE-1 inhibitor design using pharmacophore modeling, in silico screening and molecular docking studies. BMC Bioinformatics 12(Suppl 1):S28, 2011.
Kacker, P., G. Bottegoni, and A. Cavalli. Computational methods in the discovery and design of BACE-1 inhibitors. Curr. Med. Chem. 19:6095–6111, 2012.
Karubiu, W., S. Bhakat, and M. S. Soliman. Compensatory role of double mutation N348I/M184V on nevirapine binding landscape: insight from molecular dynamics simulation. Protein J. 33:1–15, 2014.
Kitchen, D. B., H. Decornez, J. R. Furr, and J. Bajorath. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat. Rev. Drug Dis. 3:935–949, 2004.
Kumalo, H. M., S. Bhakat, and M. E. Soliman. Investigation of flap flexibility of beta-secretase using molecular dynamic simulations. J. Biomol. Struct. Dyn., 2015. doi:10.1080/07391102.2015.106483.
Kumar, A., S. Roy, S. Tripathi, and A. Sharma. Molecular docking based virtual screening of natural compounds as potential BACE1 inhibitors: 3D QSAR pharmacophore mapping and molecular dynamics analysis. J. Biomol. Struct. Dyn., 2015. doi:10.1080/07391102.2015.1022603.
Kumar, V., S. Krishna, and M. I. Siddiqi. Virtual screening strategies: recent advances in the identification and design of anti-cancer agents. Methods 71:64–70, 2015.
Kumar, A., A. Singh, and Ekavali. A review on Alzheimer’s disease pathophysiology and its management: an update. Pharmacol. Rep. 67:195–203, 2015.
Lipinski, C. A., F. Lombardo, B. W. Dominy, and P. J. Feeney. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 46:3–26, 2001.
Machauer, R., K. Laumen, S. Veenstra, J.-M. Rondeau, M. Tintelnot-Blomley, C. Betschart, A.-L. Jaton, S. Desrayaud, M. Staufenbiel, S. Rabe, P. Paganetti, and U. Neumann. Macrocyclic peptidomimetic β-secretase (BACE-1) inhibitors with activity in vivo. Bioorg. Med. Chem. Lett. 19:1366–1370, 2009.
Machauer, R., S. Veenstra, J.-M. Rondeau, M. Tintelnot-Blomley, C. Betschart, U. Neumann, and P. Paganetti. Structure-based design and synthesis of macrocyclic peptidomimetic β-secretase (BACE-1) inhibitors. Bioorg. Med. Chem. Lett. 19:1361–1365, 2009.
Martin, K. R., P. Narang, J. L. Medina-Franco, N. Meurice, and J. P. MacKeigan. Integrating virtual and biochemical screening for protein tyrosine phosphatase inhibitor discovery. Methods 65:219–228, 2014.
Martinez, L. Automatic identification of mobile and rigid substructures in molecular dynamics simulations and fractional structural fluctuation analysis. PloS one 10:e0119264, 2015.
Menting, K. W., and J. A. Claassen. β-secretase inhibitor; a promising novel therapeutic drug in Alzheimer’s disease. Front. Aging Neurosci. 6:165, 2014. doi:10.3389/fnagi.2014.00165
Mohamed, T., J. C. Yeung, M. S. Vasefi, M. A. Beazely, and P. P. Rao. Development and evaluation of multifunctional agents for potential treatment of Alzheimer’s disease: application to a pyrimidine-2,4-diamine template. Bioorg. Med. Chem. Lett. 22:4707–4712, 2012.
Morris, G. M., D. S. Goodsell, R. S. Halliday, R. Huey, W. E. Hart, R. K. Belew, and A. J. Olson. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 19:1639–1662, 1998.
Morris, G. M., R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell, and A. J. Olson. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30:2785–2791, 2009.
Muegge, I., D. Collin, B. Cook, M. Hill-Drzewi, J. Horan, S. Kugler, M. Labadia, X. Li, L. Smith, and Y. Zhang. Discovery of 1,3-dihydro-2,1,3-benzothiadiazole 2,2-dioxide analogs as new RORC modulators. Bioorg. Med. Chem. Lett. 25:1892–1895, 2015.
Nagarajan, S., M. J. Choi, Y. S. Cho, S. J. Min, G. Keum, S. J. Kim, C. S. Lee, and A. N. Pae. Tubulin inhibitor identification by bioactive conformation alignment pharmacophore (BCAP)-guided virtual screening. Chem. Biol. Drug Des. 2015. doi:10.1111/cbdd.12568.
Oehlrich, D., H. Prokopcova, and H. J. Gijsen. The evolution of amidine-based brain penetrant BACE1 inhibitors. Bioorg. Med. Chem. Lett. 24:2033–2045, 2014.
Ohno, M. Roles of eIF2alpha kinases in the pathogenesis of Alzheimer’s disease. Front. Mol. Neurosci. 7:22, 2014.
Pautasso, C., R. Troia, M. Genuardi, and A. Palumbo. Pharmacophore modeling technique applied for the discovery of proteasome inhibitors. Expert Opin. Drug Discov. 9:931–943, 2014.
Poredos, P., D. Celan, J. Mozina, and M. Jezersek. Determination of the human spine curve based on laser triangulation. BMC Med. Imag. 15:2, 2015.
Razzaghi-Asl, N., O. Firuzi, B. Hemmateenejad, K. Javidnia, N. Edraki, and R. Miri. Design and synthesis of novel 3,5-bis-N-(aryl/heteroaryl) carbamoyl-4-aryl-1,4-dihydropyridines as small molecule BACE-1 inhibitors. Bioorg. Med. Chem. 21:6893–6909, 2013.
Rojo, I., J. A. Martín, H. Broughton, D. Timm, J. Erickson, H.-C. Yang, and J. R. McCarthy. Macrocyclic peptidomimetic inhibitors of β-secretase (BACE): first X-ray structure of a macrocyclic peptidomimetic-BACE complex. Bioorg. Med. Chem. Lett. 16:191–195, 2006.
Sabbagh, J. J., J. W. Kinney, and J. L. Cummings. Animal systems in the development of treatments for Alzheimer’s disease: challenges, methods, and implications. Neurobiol. Aging 34:169–183, 2013.
Sanner, M. F. Python: a programming language for software integration and development. J. Mol. Graph. Model. 17:57–61, 1999.
Savonenko, A. V., T. Melnikova, T. Li, D. L. Price, and P. C. Wong. Chapter 21—Alzheimer disease. In: Neurobiology of Brain Disorders, edited by M. J. Z. P. R. T. Coyle. San Diego: Academic Press, 2015, pp. 321–338.
Semighini, E. P. In silico design of beta-secretase inhibitors in Alzheimer’s disease. Chem. Biol. Drug Des. 86:284–290, 2014.
Shimmyo, Y., T. Kihara, A. Akaike, T. Niidome, and H. Sugimoto. Flavonols and flavones as BACE-1 inhibitors: Structure–activity relationship in cell-free, cell-based and in silico studies reveal novel pharmacophore features. Biochim. Biophys. Acta 1780:819–825, 2008.
Silva, T., J. Reis, J. Teixeira, and F. Borges. Alzheimer’s disease, enzyme targets and drug discovery struggles: from natural products to drug prototypes. Ageing Res. Rev. 15:116–145, 2014.
Sindhu, T., and P. Srinivasan. Identification of potential dual agonists of FXR and TGR5 using e-pharmacophore based virtual screening. Mol. Biosyst. 11:1305–1318, 2015.
Soliman, M. E. S. A hybrid structure/pharmacophore-based virtual screening approach to design potential leads: a computer-aided design of South African HIV-1 subtype c protease inhibitors. Drug Dev. Res. 74:283–295, 2013.
Tang, J., and G. Koelsch. Chapter 14—memapsin 2. In: Handbook of Proteolytic Enzymes, edited by N. D. R. Salvesen. London: Academic Press, 2013, pp. 87–93.
Trott, O., and A. J. Olson. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31:455–461, 2010.
Vassar, R. BACE1: the beta-secretase enzyme in Alzheimer’s disease. J. Mol. Neurosci. 23:105–114, 2004.
Wyss, D. F., Y. S. Wang, H. L. Eaton, C. Strickland, J. H. Voigt, Z. Zhu, and A. W. Stamford. Combining NMR and X-ray crystallography in fragment-based drug discovery: discovery of highly potent and selective BACE-1 inhibitors. Top. Curr. Chem. 317:83–114, 2012.
Xu, W., G. Chen, W. Zhu, and Z. Zuo. Molecular docking and structure–activity relationship studies on benzothiazole based non-peptidic BACE-1 inhibitors. Bioorg. Med. Chem. Lett. 20:6203–6207, 2010.
Xu, Y., M. J. Li, H. Greenblatt, W. Chen, A. Paz, O. Dym, Y. Peleg, T. Chen, X. Shen, J. He, H. Jiang, I. Silman, and J. L. Sussman. Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations. Acta Crystallogr. D Biol. Crystallogr. 68:13–25, 2012.
Forli, S. AutoDock | Raccoon: an automated tool for preparing AutoDock virtual screenings.
Yan, X. X., C. Ma, W. P. Gai, H. Cai, and X. G. Luo. Can BACE1 inhibition mitigate early axonal pathology in neurological diseases? J. Alzheimer’s Dis. 38:705–718, 2014.
Yan, R., and R. Vassar. Targeting the beta secretase BACE1 for Alzheimer’s disease therapy. Lancet Neurol. 13:319–329, 2014.
Yi Mok, N., J. Chadwick, K. A. B. Kellett, N. M. Hooper, A. P. Johnson, and C. W. G. Fishwick. Discovery of novel non-peptide inhibitors of BACE-1 using virtual high-throughput screening. Bioorg. Med. Chem. Lett. 19:6770–6774, 2009.
Zhang, Z., N. Guan, T. Li, D. E. Mais, and M. Wang. Quality control of cell-based high-throughput drug screening. Acta Pharm. Sin. B 2:429–438, 2012.
Zhou, Z. G., Y. L. Wang, and S. H. Bryant. Computational analysis of the Cathepsin B inhibitors activities through LR-MMPBSA binding affinity calculation based on docked complex. J. Comput. Chem. 30:2165–2175, 2009.
Zhu, Y. P., K. Xiao, H. P. Yu, L. P. Ma, B. Xiong, H. Y. Zhang, X. Wang, J. Y. Li, J. Li, and J. K. Shen. Discovery of potent beta-secretase (bace-1) inhibitors by the synthesis of isophthalamide-containing hybrids. Acta Pharmacol. Sin. 30:259–269, 2009.
Acknowledgement
Authors like to acknowledge NRF, South Africa and School of Health Sciences, University of KwaZulu-Natal, Westville for financial support and cBio cluster at MSKCC for computational resources.
Conflict of Interest
Kumalo HM and Mahmoud E. Soliman declare that they have no conflict of interest.
Ethical standards
No animal or human studies were carried out by the authors for this article.
Author information
Authors and Affiliations
Corresponding author
Additional information
Associate Editor Yiannis Kaznessis oversaw the review of this article.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kumalo, H.M., Soliman, M.E. Per-Residue Energy Footprints-Based Pharmacophore Modeling as an Enhanced In Silico Approach in Drug Discovery: A Case Study on the Identification of Novel β-Secretase1 (BACE1) Inhibitors as Anti-Alzheimer Agents. Cel. Mol. Bioeng. 9, 175–189 (2016). https://doi.org/10.1007/s12195-015-0421-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12195-015-0421-8