Abstract
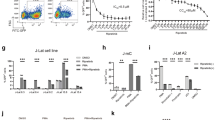
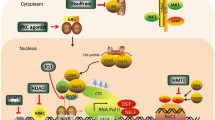
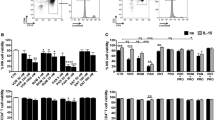
Human immunodeficiency virus 1 (HIV) latency remains a significant obstacle to curing infected patients. One promising therapeutic strategy is to purge the latent cellular reservoir by activating latent HIV with latency-reversing agents (LRAs). In some cases, co-drugging with multiple LRAs is necessary to activate latent infections, but few studies have established quantitative criteria for determining when co-drugging is required. Here we systematically quantified drug interactions between histone deacetylase inhibitors and transcriptional activators of HIV and found that the need for co-drugging is determined by the proximity of latent infections to the chromatin-regulated viral gene activation threshold at the viral promoter. Our results suggest two classes of latent viral integrations: those far from the activation threshold that benefit from co-drugging, and those close to the threshold that are efficiently activated by a single drug. Using a primary T cell model of latency, we further demonstrated that the requirement for co-drugging was donor dependent, suggesting that the host may set the level of repression of latent infections. Finally, we showed that single drug or co-drugging doses could be optimized, via repeat stimulations, to minimize unwanted side effects while maintaining robust viral activation. Our results motivate further study of patient-specific latency-reversing strategies.







Similar content being viewed by others
References
Archin, N. M., A. L. Liberty, A. D. Kashuba, S. K. Choudhary, J. D. Kuruc, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 487:482–485, 2012.
Barboric, M., J. H. N. Yik, N. Czudnochowski, Z. Yang, R. Chen, et al. Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription. Nucleic Acids Res. 35:2003–2012, 2007.
Blazkova, J., K. Trejbalova, F. Gondois-Rey, P. Halfon, P. Philibert, et al. CpG Methylation Controls Reactivation of HIV from Latency. PLoS Pathog. 5:e1000554, 2009.
Bliss, C. I. The calculation of microbial assays. Bacteriol. Rev. 20:243–258, 1956.
Böhnlein, E., J. W. Lowenthal, M. Siekevitz, D. W. Ballard, B. R. Franza, et al. The same inducible nuclear proteins regulates mitogen activation of both the interleukin-2 receptor-alpha gene and type 1 HIV. Cell 53:827–836, 1988.
Bosque, A., and V. Planelles. Induction of HIV-1 latency and reactivation in primary memory CD4+ T cells. Blood 113:58–65, 2009.
Bosque, A., and V. Planelles. Studies of HIV-1 latency in an ex vivo model that uses primary central memory T cells. Methods 53:54–61, 2011.
Bullen, C. K., G. M. Laird, C. M. Durand, J. D. Siliciano, and R. F. Siliciano. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat. Med. 20:425–429, 2014.
Burnett, J. C., K.-I. Lim, A. Calafi, J. J. Rossi, D. V. Schaffer, et al. Combinatorial latency reactivation for HIV-1 subtypes and variants. J. Virol. 84:5958–5974, 2010.
Burnett, J. C., K. Miller-Jensen, P. S. Shah, A. P. Arkin, and D. V. Schaffer. Control of stochastic gene expression by host factors at the HIV promoter. PLoS Pathog. 5:e1000260, 2009.
Choudhary, S. K., N. M. Archin, and D. M. Margolis. Hexamethylbisacetamide and disruption of human immunodeficiency virus type 1 latency in CD4(+) T cells. J. Infect. Dis. 197:1162–1170, 2008.
Contreras, X., M. Barboric, T. Lenasi, and B. M. Peterlin. HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog. 3:1459–1469, 2007.
Contreras, X., M. Schweneker, C. S. Chen, J. M. McCune, S. G. Deeks, et al. Suberoylanilide hydroxamic acid reactivates HIV from latently infected cells. J. Biol. Chem. 284:6782–6789, 2009.
Coull, J. J., F. Romerio, J. M. Sun, J. L. Volker, K. M. Galvin, et al. The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. J. Virol. 74:6790–6799, 2000.
Duh, E. J., W. J. Maury, T. M. Folks, A. S. Fauci, and A. B. Rabson. Tumor necrosis factor alpha activates human immunodeficiency virus type 1 through induction of nuclear factor binding to the NF-kappa B sites in the long terminal repeat. Proc Natl Acad Sci USA 86:5974–5978, 1989.
Durand, C. M., J. N. Blankson, and R. F. Siliciano. Developing strategies for HIV-1 eradication. Trends Immunol. 33:554–562, 2012.
Emiliani, S., C. Van Lint, W. Fischle, P. Paras, Jr., M. Ott, et al. A point mutation in the HIV-1 Tat responsive element is associated with postintegration latency. Proc. Natl. Acad. Sci. USA 93:6377–6381, 1996.
Fernandez, G., and S. L. Zeichner. Cell line-dependent variability in HIV activation employing DNMT inhibitors. Virol. J. 7:266, 2010.
Finzi, D., J. Blankson, J. D. Siliciano, J. B. Margolick, K. Chadwick, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512–517, 1999.
Finzi, D., M. Hermankova, T. Pierson, L. M. Carruth, C. Buck, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300, 1997.
Fitzgerald, J. B., B. Schoeberl, U. B. Nielsen, and P. K. Sorger. Systems biology and combination therapy in the quest for clinical efficacy. Nat. Chem. Biol. 2:458–466, 2006.
Ganesh, L., E. Burstein, A. Guha-Niyogi, M. K. Louder, J. R. Mascola, et al. The gene product Murr1 restricts HIV-1 replication in resting CD4+ lymphocytes. Nature 426:853–857, 2003.
Han, Y., K. Lassen, D. Monie, A. R. Sedaghat, S. Shimoji, et al. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J. Virol. 78:6122–6133, 2004.
Ho, Y. C., L. Shan, N. N. Hosmane, J. Wang, S. B. Laskey, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155:540–551, 2013.
Jiang, G., A. Espeseth, D. J. Hazuda, and D. M. Margolis. c-Myc and Sp1 contribute to proviral latency by recruiting histone deacetylase 1 to the human immunodeficiency virus type 1 promoter. J. Virol. 81:10914–10923, 2007.
Jordan, A., D. Bisgrove, and E. Verdin. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 22:1868–1877, 2003.
Kao, S. Y., A. F. Calman, P. A. Luciw, and B. M. Peterlin. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 330:489–493, 1987.
Kasowski, M., S. Kyriazopoulou-Panagiotopoulou, F. Grubert, J. B. Zaugg, A. Kundaje, et al. Extensive variation in chromatin states across humans. Science 342:750–752, 2013.
Kauder, S. E., A. Bosque, A. Lindqvist, V. Planelles, and E. Verdin. Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog. 5:e1000495, 2009.
Kim, Y. K., C. F. Bourgeois, R. Pearson, M. Tyagi, M. J. West, et al. Recruitment of TFIIH to the HIV LTR is a rate-limiting step in the emergence of HIV from latency. EMBO J. 25:1–9, 2006.
Kinoshita, S., L. Su, M. Amano, L. A. Timmerman, H. Kaneshima, et al. The T cell activation factor NF-ATc positively regulates HIV-1 replication and gene expression in T cells. Immunity 6:235–244, 1997.
Miller-Jensen, K., S. S. Dey, N. Pham, J. E. Foley, A. P. Arkin, et al. Chromatin accessibility at the HIV LTR promoter sets a threshold for NF-kappaB mediated viral gene expression. Integr. Biol. (Camb.) 4:661–671, 2012.
Miller-Jensen, K., R. Skupsky, P. S. Shah, A. P. Arkin, and D. V. Schaffer. Genetic selection for context-dependent stochastic phenotypes: Sp1 and TATA mutations increase phenotypic noise in HIV-1 gene expression. PLoS Comput. Biol. 9:e1003135, 2013.
Nabel, G., and D. Baltimore. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 326:711–713, 1987.
Ott, M., M. Geyer, and Q. Zhou. The control of HIV transcription: keeping RNA polymerase II on track. Cell Host Microbe 10:426–435, 2011.
Quivy, V., E. Adam, Y. Collette, D. Demonte, A. Chariot, et al. Synergistic activation of human immunodeficiency virus type 1 promoter activity by NF-kappaB and inhibitors of deacetylases: potential perspectives for the development of therapeutic strategies. J. Virol. 76:11091–11103, 2002.
Reuse, S., M. Calao, K. Kabeya, A. Guiguen, J.-S. Gatot, et al. Synergistic activation of HIV-1 expression by deacetylase inhibitors and prostratin: implications for treatment of latent infection. PLoS ONE 4:e6093, 2009.
Richman, D. D., D. M. Margolis, M. Delaney, W. C. Greene, D. Hazuda, et al. The challenge of finding a cure for HIV infection. Science 323:1304–1307, 2009.
Rowinsky, E. K., D. S. Ettinger, L. B. Grochow, R. B. Brundrett, A. E. Cates, et al. Phase I and pharmacologic study of hexamethylene bisacetamide in patients with advanced cancer. J. Clin. Oncol. 4:1835–1844, 1986.
Selby, M. J., and B. M. Peterlin. Trans-activation by HIV-1 Tat via a heterologous RNA binding protein. Cell 62:769–776, 1990.
Tyagi, M., and J. Karn. CBF-1 promotes transcriptional silencing during the establishment of HIV-1 latency. EMBO J. 26:4985–4995, 2007.
Tyagi, M., R. J. Pearson, and J. Karn. Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and P-TEFb restriction. J. Virol. 84:6425–6437, 2010.
Weinberger, L. S., J. C. Burnett, J. E. Toettcher, A. P. Arkin, and D. V. Schaffer. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell 122:169–182, 2005.
Williams, S. A., L.-F. Chen, H. Kwon, C. M. Ruiz-Jarabo, E. Verdin, et al. NF-κB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J. 25:139–149, 2006.
Acknowledgments
This work was supported by the Bill and Melinda Gates Foundation’s Grand Challenges Explorations (Round 7 to K.M.J.) and the National Science Foundation (NSF CBET-1264246 to K.M.J.). V.C.W. was supported by a National Institutes of Health Predoctoral Training Grant in Genetics (2T32GM007499-36, 5T32GM007499-34, 5T32GM007499-35).
Conflict of interest
V.C.W., L.E.F., N.M.A., Q.X., S.S.D., and K.M.J. declare that they have no conflicts of interest.
Ethical Standards
No human studies or animal studies were carried out by the authors for this article.
Author information
Authors and Affiliations
Corresponding author
Additional information
Associate Editor David Schaffer oversaw the review of this article.
This paper is part of the 2014 Young Innovators Issue.
Kathryn Miller-Jensen is an Assistant Professor of Biomedical Engineering and Molecular, Cellular, and Developmental Biology at Yale University. Her lab uses experimental and computational approaches to study signaling and transcriptional regulation in response to pathogens, with a focus on activation of latent HIV, as well as innate immune signaling. Her work is funded by the National Institutes of Health, the National Science Foundation, and the Bill and Melinda Gates Foundation. Kathryn was an NIH NSRA Postdoctoral Fellow at the University of California at Berkeley and she holds a Ph.D. in Chemical Engineering from the Massachusetts Institute of Technology and A.B. and B.E. degrees in Engineering Sciences from Dartmouth College. She is a member of the Biomedical Engineering Society and a former Christine Mirzayan Science and Technology Policy Fellow at the National Academies in Washington, DC.

Victor C. Wong and Linda E. Fong have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wong, V.C., Fong, L.E., Adams, N.M. et al. Quantitative Evaluation and Optimization of Co-drugging to Improve Anti-HIV Latency Therapy. Cel. Mol. Bioeng. 7, 320–333 (2014). https://doi.org/10.1007/s12195-014-0336-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12195-014-0336-9