Abstract
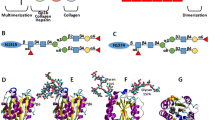
The three tandem A domains (A1, A2, and A3) of von Willebrand factor (VWF) play critical roles for its functions. The A1 and A3 domains contain respective binding sites for platelet glycoprotein Ib (GPIb) and collagen. The A2 domain hosts a proteolytic site for the VWF-cleavage enzyme A Disintegrin And Metalloprotease with a ThromboSpondin type 1 motifs 13 (ADAMTS-13). Previous studies suggested that shear flow assists the ADAMTS-13 cleavage of VWF by unfolding the A2 domain and thus exposing the cryptic proteolytic site. Here we used steered molecular dynamics (SMD) to simulate the unfolding of the A1 and A2 domains by tensile force. The forced unfolding of A2 started from the C-terminus because of its specific topology. The β-strands of A2 were pulled out sequentially, generating sawtooth-like peaks in the force-extension curves. The disulfide bond between A1 N- and C-termini prevented it from unfolding. After eliminating the disulfide bond, A1 was unfolded similarly as A2 in terms of the β-strand pullouts, but differed in the unfolding of helices. The major resistance of A1 and A2 to unfolding came from the hydrogen bond networks of the central β-sheets. Two different unfolding pathways of the β-strands were observed, where the sliding pathway encountered much higher energy barrier than the unzipping pathway.










Similar content being viewed by others
References
Arya, M., B. Anvari, G.M. Romo, M.A. Cruz, J.F. Dong, L.V. McIntire, J.L. Moake, and J.A. Lopez. Ultralarge multimers of von Willebrand factor form spontaneous high-strength bonds with the platelet glycoprotein Ib-IX complex: studies using optical tweezers. Blood. 99(11): 3971–3977, 2002.
Auton, M., M.A. Cruz, and J. Moake. Conformational stability and domain unfolding of the von Willebrand factor A domains. J Mol Biol. 366(3): 986–1000, 2007.
Bienkowska, J., M. Cruz, A. Atiemo, R. Handin, and R. Liddington. The von Willebrand factor A3 domain does not contain a metal ion-dependent adhesion site motif. J Biol Chem. 272(40): 25162–25167, 1997.
Bond, C.S. TopDraw: a sketchpad for protein structure topology cartoons. Bioinformatics. 19(2): 311–312, 2003.
Brahms, S. and J. Brahms. Determination of protein secondary structure in solution by vacuum ultraviolet circular dichroism. J Mol Biol. 138(2): 149–178, 1980.
Carl, P., C.H. Kwok, G. Manderson, D.W. Speicher, and D.E. Discher. Forced unfolding modulated by disulfide bonds in the Ig domains of a cell adhesion molecule. Proc Natl Acad Sci U S A. 98(4): 1565–1570, 2001.
Case, D.A., T. A. Darden, T. E. Cheatham, III, C. L. Simmerling, J. Wang, R. E. Duke, R. Luo, K. M. Merz, B. Wang, D. A. Pearlman, M. Crowley, S. Brozell, V. Tsui, H. Gohlke, J. Mongan, V. Hornak, G. Cui, P. Beroza, C. Schafmeister, J. W. Caldwell, W. S. Ross, and P. A. Kollman. AMBER 8. University of California, San Francisco, 2004.
Castaman, G., A.B. Federici, F. Rodeghiero, and P.M. Mannucci. von Willebrand’s disease in the year 2003: towards the complete identification of gene defects for correct diagnosis and treatment. Haematologica. 88(1): 94–108, 2003.
Dent, J.A., S.D. Berkowitz, J. Ware, C.K. Kasper, and Z.M. Ruggeri. Identification of a cleavage site directing the immunochemical detection of molecular abnormalities in type IIA von Willebrand factor. Proc Natl Acad Sci U S A. 87(16): 6306–6310, 1990.
Dong, J.F., J.L. Moake, L. Nolasco, A. Bernardo, W. Arceneaux, C.N. Shrimpton, A.J. Schade, L.V. McIntire, K. Fujikawa, and J.A. Lopez. ADAMTS-13 rapidly cleaves newly secreted ultralarge von Willebrand factor multimers on the endothelial surface under flowing conditions. Blood. 100(12): 4033–4039, 2002.
Duan, Y., C. Wu, S. Chowdhury, M.C. Lee, G. Xiong, W. Zhang, R. Yang, P. Cieplak, R. Luo, T. Lee, J. Caldwell, J. Wang, and P. Kollman. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 24(16): 1999–2012, 2003.
Emsley, J., M. Cruz, R. Handin, and R. Liddington. Crystal structure of the von Willebrand factor A1 domain and implications for the binding of platelet glycoprotein Ib. J Biol Chem. 273(17): 10396–10401, 1998.
Franchini, M. and G. Lippi. von Willebrand factor and thrombosis. Ann Hematol. 85(7): 415–423, 2006.
Frishman, D. and P. Argos. Knowledge-based protein secondary structure assignment. Proteins. 23(4): 566–579, 1995.
Gerhardt, S., G. Hassall, P. Hawtin, E. McCall, L. Flavell, C. Minshull, D. Hargreaves, A. Ting, R.A. Pauptit, A.E. Parker, and W.M. Abbott. Crystal structures of human ADAMTS-1 reveal a conserved catalytic domain and a disintegrin-like domain with a fold homologous to cysteine-rich domains. J Mol Biol. 373(4): 891–902, 2007.
Huizinga, E.G., R. Martijn van der Plas, J. Kroon, J.J. Sixma, and P. Gros. Crystal structure of the A3 domain of human von Willebrand factor: implications for collagen binding. Structure. 5(9): 1147–1156, 1997.
Huizinga, E.G., S. Tsuji, R.A. Romijn, M.E. Schiphorst, P.G. de Groot, J.J. Sixma, and P. Gros. Structures of glycoprotein Ibalpha and its complex with von Willebrand factor A1 domain. Science. 297(5584): 1176–1179, 2002.
Humphrey, W., A. Dalke, and K. Schulten. VMD: visual molecular dynamics. J. Mol. Graph. 14(1): 33–38, 27–38, 1996.
Lee, J.O., P. Rieu, M.A. Arnaout, and R. Liddington. Crystal structure of the A domain from the alpha subunit of integrin CR3 (CD11b/CD18). Cell. 80(4): 631–638, 1995.
Levy, G.G., W.C. Nichols, E.C. Lian, T. Foroud, J.N. McClintick, B.M. McGee, A.Y. Yang, D.R. Siemieniak, K.R. Stark, R. Gruppo, R. Sarode, S.B. Shurin, V. Chandrasekaran, S.P. Stabler, H. Sabio, E.E. Bouhassira, J.D. Upshaw, Jr., D. Ginsburg, and H.M. Tsai. Mutations in a member of the ADAMTS gene family cause thrombotic thrombocytopenic purpura. Nature. 413(6855): 488–494, 2001.
Matsushita, T. and J.E. Sadler. Identification of amino acid residues essential for von Willebrand factor binding to platelet glycoprotein Ib. Charged-to-alanine scanning mutagenesis of the A1 domain of human von Willebrand factor. J Biol Chem. 270(22): 13406–13414, 1995.
Mazzucato, M., P. Spessotto, A. Masotti, L. De Appollonia, M.R. Cozzi, A. Yoshioka, R. Perris, A. Colombatti, and L. De Marco. Identification of domains responsible for von Willebrand factor type VI collagen interaction mediating platelet adhesion under high flow. J Biol Chem. 274(5): 3033–3041, 1999.
Michiels, J.J., A. Gadisseur, U. Budde, Z. Berneman, M. van der Planken, W. Schroyens, A. van de Velde, and H. van Vliet. Characterization, classification, and treatment of von Willebrand diseases: a critical appraisal of the literature and personal experiences. Semin Thromb Hemost. 31(5): 577–601, 2005.
Mosyak, L., K. Georgiadis, T. Shane, K. Svenson, T. Hebert, T. McDonagh, S. Mackie, S. Olland, L. Lin, X. Zhong, R. Kriz, E.L. Reifenberg, L.A. Collins-Racie, C. Corcoran, B. Freeman, R. Zollner, T. Marvell, M. Vera, P.E. Sum, E.R. Lavallie, M. Stahl, and W. Somers. Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5. Protein Sci. 17(1): 16–21, 2008.
Padilla, A., J.L. Moake, A. Bernardo, C. Ball, Y. Wang, M. Arya, L. Nolasco, N. Turner, M.C. Berndt, B. Anvari, J.A. Lopez, and J.F. Dong. P-selectin anchors newly released ultralarge von Willebrand factor multimers to the endothelial cell surface. Blood. 103(6): 2150–2156, 2004.
Phillips, J.C., R. Braun, W. Wang, J. Gumbart, E. Tajkhorshid, E. Villa, C. Chipot, R.D. Skeel, L. Kale, and K. Schulten. Scalable molecular dynamics with NAMD. J Comput Chem. 26(16): 1781–1802, 2005.
Qu, A. and D.J. Leahy. Crystal structure of the I-domain from the CD11a/CD18 (LFA-1, alpha L beta 2) integrin. Proc Natl Acad Sci U S A. 92(22): 10277–10281, 1995.
Romijn, R.A., B. Bouma, W. Wuyster, P. Gros, J. Kroon, J.J. Sixma, and E.G. Huizinga. Identification of the collagen-binding site of the von Willebrand factor A3-domain. J Biol Chem. 276(13): 9985–9991, 2001.
Sadler, J.E. von Willebrand factor: two sides of a coin. J Thromb Haemost. 3(8): 1702–1709, 2005.
Shieh, H.S., K.J. Mathis, J.M. Williams, R.L. Hills, J.F. Wiese, T.E. Benson, J.R. Kiefer, M.H. Marino, J.N. Carroll, J.W. Leone, A.M. Malfait, E.C. Arner, M.D. Tortorella, and A. Tomasselli. High resolution crystal structure of the catalytic domain of ADAMTS-5 (aggrecanase-2). J Biol Chem. 283(3): 1501–1507, 2008.
Sutherland, J.J., L.A. O’Brien, D. Lillicrap, and D.F. Weaver. Molecular modeling of the von Willebrand factor A2 Domain and the effects of associated type 2A von Willebrand disease mutations. J Mol Model. 10(4): 259–270, 2004.
Tsai, H.M. Deficiency of ADAMTS13 and thrombotic thrombocytopenic purpura. Blood. 100(10): 3839–3840; author reply 3840–3832, 2002.
Tsai, H.M., Sussman, II, and R.L. Nagel. Shear stress enhances the proteolysis of von Willebrand factor in normal plasma. Blood. 83(8): 2171–2179, 1994.
Wiita, A.P., S.R. Ainavarapu, H.H. Huang, and J.M. Fernandez. Force-dependent chemical kinetics of disulfide bond reduction observed with single-molecule techniques. Proc Natl Acad Sci U S A. 103(19): 7222–7227, 2006.
Wiita, A.P., R. Perez-Jimenez, K.A. Walther, F. Grater, B.J. Berne, A. Holmgren, J.M. Sanchez-Ruiz, and J.M. Fernandez. Probing the chemistry of thioredoxin catalysis with force. Nature. 450(7166): 124–127, 2007.
Yago, T., J. Lou, T. Wu, J. Yang, J.J. Miner, L. Coburn, J.A. Lopez, M.A. Cruz, J.F. Dong, L.V. McIntire, R.P. McEver, and C. Zhu. Platelet glycoprotein Ibalpha forms catch bonds with human WT vWF but not with type 2B von Willebrand disease vWF. J Clin Invest. 118(9): 3195–3207, 2008.
Acknowledgments
We thank Dr. Stephen Harvey for kindly providing computational resources for the MD simulations. We are also grateful for supercomputer time provided by TeraGrid via NCSA DAC grant MCB080011N and LRAC grant MCA08X014. This work is supported by NIH grant HL091020 (C.Z.) and a Scientist Development Grant from American Heart Association (J.L.).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Video 1
A2 was pulled at the C-terminal Cα atom while the N-terminal Cα atom was fixed. α-helices are shown as coiled ribbons, β-strands as ribbons with arrows, and loops as tubes. A blue and a red sphere indicate, respectively, the N- and C-terminal Cα atoms. The backbone atoms of Tyr1605 and Met1606 adjacent to the proteolytic site are shown as green spheres. (AVI 13994 kb)
Video 2
A2 was pulled at the N-terminal Cα atom while the C-terminal Cα atom was fixed. To facilitate comparison with Video 1, A2 in each frame has been translated such that the N-terminal Cα atom stays at its starting position to allow the structured portion of A2 to remain in view. The representations of A2 are the same as in Video 1. (AVI 17066 kb)
A1 with an intact disulfide bond was pulled at the C-terminal Cα atom while the N-terminal Cα atom was fixed. α-helices are shown as coiled ribbons, β-strands as ribbons with arrows, and loops as tubes. A blue and a red sphere indicate, respectively, the N- and C-terminal Cα atoms. The disulfide bond atoms are shown as bonds. (AVI 2814 kb)
Video 4
A1 with a broken disulfide bond was pulled at the C-terminal Cα atom while the N-terminal Cα atom was fixed. The representations of A1 are the same as in Video 3. (AVI 13738 kb)
Rights and permissions
About this article
Cite this article
Chen, W., Lou, J. & Zhu, C. Molecular Dynamics Simulated Unfolding of von Willebrand Factor A Domains by Force. Cel. Mol. Bioeng. 2, 75–86 (2009). https://doi.org/10.1007/s12195-009-0051-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12195-009-0051-0