Abstract
The goal of this study was to examine the possibility of verifying the origin and authenticity of Polish cheese, Oscypek marked with protected designation of origin (PDO) label, based on its volatile profile. Oscypek and its false imitations Oscypek-like cheeses were analyzed using a solid-phase microextraction-mass spectrometry method (SPME-MS), which involved volatiles extraction by SPME with subsequent volatiles characterization by MS without particular components separation by gas chromatography. Collected data were treated using principal component analysis (PCA), linear discriminant analysis (LDA), soft independent modeling of class analogy (SIMCA), and support vector machines (SVM). The results of this study demonstrated that SPME-MS method combined with chemometric tools can be successfully used as electronic nose and applied to detect mislabeling and adulteration of PDO Oscypek cheese.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The popularity of protected designation of origin (PDO) products is still growing; their chemical, microbial, and sensory characterization is crucial to differentiate them from similar products without PDO label (Mallia et al. 2005), to provide final consumer’s satisfaction and protect from adulteration. Unfortunately, PDO products, alike other food products, are subjected to fraud. The most common adulteration among food producers is the partial or complete substitution of authentic material, easily available, and cheaper (Hrbek et al. 2014) and using advanced technology. The authenticity of Oscypek PDO (Commission Regulation (EC) 2006) is associated with such factors as a geographical area of production (Podhale Region of Polish Tatra Mountains), materials (raw ewe’s milk), and traditional technology. Raw ewe’s milk should be obtained from a particular breed of mountain sheep called Polish Highland, but the addition of cow’s milk is also allowed, only if the milk comes from a specific mountain breed called Polish Red, and the addition is only up to 40 %. Additionally, traditional technology based on hand processing, application of wooden tools, and natural preservation methods such as brining and smoking in shepherds’ huts must be employed (Majcher et al. 2011; Majcher and Jeleń 2011). Such process is typical of a defined product, and therefore, the resulting volatile composition could be an interesting indicator of Oscypek origin. However, to resolve authenticity issues, unique markers or group of markers should be identified (Guerreiro et al. 2013). In dairy products, proteins have been widely used as adulteration markers with different immunological, chromatographic, and electrophoresis techniques employed (Mafra et al. 2008). Though, besides proteins, volatile compounds identified by chromatographic methods and electronic nose could be used for the determination of geographic origin of some dairy products (Pillonel et al. 2003).
Polish PDO cheese Oscypek has a distinct characteristic aroma described previously by authors (Majcher and Jeleń 2011). Flavor is recognized as one of the key attribute that determines cheese quality (Adda 1986). It has been reported also that cheese aroma is influenced by animal feeding, milk origin, or cheese-making method (Moio et al. 1996; Urbach 1997; Barron et al. 2005). The comparison of Oscypek and Oscypek-like cheeses volatile profiles revealed that the aroma profile of Oscypek had larger amount of volatile and more developed aroma than other cheeses. Additionally, Oscypek-like cheeses produced industrially were less developed in flavor than artisan cheese (Majcher et al. 2010).
The continuously developed analytical techniques could be a useful tool in food quality and authenticity control, leading to identify and quantify trace amounts of compounds. Therefore, chromatographic methods coupled with chemometrics could be used to confirm product authenticity (Tu et al. 2014; Lv et al. 2014). Solid-phase microextraction (SPME) with combination of gas chromatography-mass spectrometry is recently the key method of choice for the analysis of volatile pattern of different food product, including cheeses (Berezińska and Obiedziński 2007; Majcher et al. 2010; Urgeghe et al. 2012). SPME-MS analysis, which is based on the average MS signal obtained from the sample without GC separation has been introduced as an electronic nose type analysis at the end of 1990s (Dittmann et al. 1998; Marsili 1999). Since then, together with chemometric tools, it was successfully applied to discriminate differences and authenticity of honey (Stanimirova et al. 2010), olive oil (Cajka et al. 2010a), beer (Cajka et al. 2010b), and raw spirits (Jeleń et al. 2010); however, there is only a few works on cheese authenticity based on volatile analysis. SPME coupled with mass spectrometry was used for rapid characterization of Camembert-type cheeses at different step of ripening (Peres et al. 2001).
Therefore, the aim of this study was to develop a rapid method for the authenticity determination of traditional Polish cheese Oscypek based on the volatile profile. Consequently, for this purpose, SPME-MS technique that combines extraction with profiling of volatile compounds without separation was used, with further treatment of the data by different statistical analysis.
Materials and Methods
Cheese Samples
Original Oscypek cheeses with PDO label were produced according to traditional technology in the Podhale Region directly from shepherd huts from Niedzica, Białka Tatrzańska, Brzegi, Bukowina Tatrzańska, and Leśnica—abbreviation “OO” (37 samples). Adulterated cheeses, Oscypek-like, were produced from cow’s milk according to traditional technology from shepherds huts—abbreviation “AO1” (36 samples). Oscypek-like cheeses were produced from ewe’s milk, according to industrial method, in a small dairy factory—abbreviation “AO2” (18 samples). Oscypek-like cheeses were produced from cow’s milk, according to industrial method, in a dairy factory—abbreviation “AO3” (22 samples). All the Oscypek-like cheeses are produced in Podhale Region. All cheese samples were stored under vacuum and frozen until analyzed. Analysis was performed within 2 months from the collection of the samples.
Sample Preparation
Frozen samples of Oscypek and Oscypek-like cheeses were grated mechanically and 10 g of samples were taken and placed in 40 mL vials, sealed with an aluminum crimp cap provided with a needle-pierceable polytetrafluoroethylene/silicone septum.
Analytical Equipment
For SPME-MS analysis, a Hewlett-Packard HP5890II coupled to a Hewlett-Packard 5971 quadrupole mass spectrometer was used. Capillary analytical column was replaced by a fused silica capillary tubing with no phase coating (fused silica, 5 m × 0.2 mm, Supelco, Bellefonte, PA).
SPME-MS Method
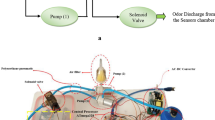
The sample vial was placed in a 50 °C heating block for 5 min to equilibrate and then the septum was pierced with the SPME needle. The fiber was exposed to the sample headspace for 15 min. After the extraction time, fiber was retracted into the needle and transferred immediately to the injection port and desorbed at 260 °C for 5 min in a splitless mode. Carboxen/PDMS (CAR/PDMS) fiber was used for extraction of volatiles, as the most effective fiber based on previously published data (Majcher et al. 2011). GC-MS operating conditions were as follows: helium flow, 0.4 mL min−1 and oven temperature, 200 °C (isothermal). The spectrometer was operated in an electron ionization (EI) mode (70 eV). The ion source was indirectly heated by a transfer line set to 280 °C. Detection was carried out in a full scan mode in a range of 33–330 m/z. All samples were run in triplicates.
Statistical Analysis
All statistical tests were performed using Statistica 10.0 software (Statsoft, Tulsa, OK) equipped with a multivariate statistics package. Principal component analysis (PCA) was used as a first step of data analysis to visualize information and to detect patterns in data, whereas linear discriminant analysis (LDA) was used to calculate classification rules for samples discrimination. Additionally, support vector machines (SVM) and soft independent modeling of class analogy (SIMCA) were also used.
Results and Discussion
As a result of SPME sampling, a single peak with an average spectrum reflecting volatile compounds adsorbed was observed (Fig. 1). The SPME-MS method employs no chromatographical separation, a single peak of unresolved volatile compounds desorbed from the fiber is obtained, and the collection of the data in mass spectrometer lasts only a few minutes. Electronic noses based on mass detection use fragment ions (m/z) which act as sensors and can be used as variables in chemometric data treatment.
Data Processing Prior to Chemometric Analysis
In the first stage, before the construction of any classification/discrimination models, the raw data (number of samples x number of variables) were logarithmically transformed and preprocessed using constant row sum approach (each variable was divided by the sum of all variables for each sample). The logarithmic transformation was used to correct significant differences in the variances obtained for the raw data. Such transformation enables an easier comparison of the sample profiles.
Because of a large volume of predictive variables and their collinearity, partial least squares (PLS) method together with variable importance in the projection (VIP) index was used to reduce the number of variables. After choosing 30 best predictors, PCA analysis were performed. First 10 components explaining over 97 % of variability were used to build classification model using linear discriminant analysis.
Principal Component Analysis
Principal component analysis (PCA) represents one of the most frequently used chemometric tool, which is applied in the first stage of the data processing. One of the most attractive features of PCA is the possibility to project particular data from a higher to a lower dimensional space and then reconstruct them without any preliminary assumptions about their distribution (Stanimirova et al. 2007). In the preliminary analysis of the different cheese data, PCA was performed to investigate clustering of samples on the basis of milk origin and technological process. The first principal component accounted for 35.4 % of variance, while the second principal component contributed 28.2 %. As Fig. 2 shows, using PCA approach, cheese samples were divided into several groups. First group described as “OO” samples is a group of Oscypek cheeses with PDO label produced according to traditional technology in Podhale Region. PCA approach let to perfectly separate ewe’s cheeses from all cheese samples. Both Oscypek-like cheeses produced from ewe’s milk, according to industrial method, in a small dairy factory “AO2” and Oscypek-like cheeses produced from cow’s milk, according to industrial method, in a dairy factory “AO3,” were ideally separated from original certified ewe’s cheeses “OO” and cheeses produced from cow’s milk according to traditional technology from shepherds hut “AO1.” Such grouping confirmed the utility of SPME-MS-PCA as a tool for the detection of adulteration of ewe’s cheeses with cow’s milk. However, this technique is less sensitive in the differentiation of industrially produced Oscypek-like cheese, both cow’s and ewe’s, samples “AO2” and “AO3” (Fig. 2).
Linear Discriminant Analysis
Linear discriminant analysis (LDA), as a supervised pattern recognition method, is probably the most frequently used. LDA is based on the determination of linear discriminant functions, which maximize the ratio of between-class variance and minimize the ratio of within-class variance, which was applied for collected data (Berrueta et al. 2007). To avoid the LDA model overfitting due to high number of variables, the first five PCs obtained with the PCA were used as the inputs for LDA. However, the classification ability using five principal components were relatively low (83 %). Only, the “OO” samples create a clearly separated group, but the discrimination among other samples is difficult (data not shown). Therefore, the LDA analysis was performed on 10 principal components. As a scatter plot of canonical values (Fig. 3) shows that the discrimination of “OO” samples is noticeable, additionally, classification groups are more cohesive than for less number of inputs. In the case of “OO” samples, the propriety of classification was 100 %, whereas for “AO1,” “AO3,” and “AO2” was 96.6, 95.5, and 83,3 %, respectively. LDA let to clearly separate ewe’s from cow’s milk cheeses, both produced with traditional technology. However, discrimination between Oscypek-like cheeses produced industrially was almost impossible. The difficulties in classification of AO3 and AO2 samples could be related not only to the relatively low number of samples in test set but also to the false producer declaration about milk origin.
Soft Independent Modeling of Class Analogy
SIMCA main idea is to build a confidence limit for each class with the help of principal component analysis and then to project the unclassified samples into each principal components space and to assign them to the class in which they fit best (Wold 1976). In SIMCA method, selection of the optimal number of principal components is a key point (Vandeginste et al. 1998). For all group samples, for a PCA model, 30 best predictors were used. On the base of classification matrix, the percentage of classification propriety was obtained. The highest percentage of correctly classified samples was in “OO” group which was 100 %. Afterwards, for “AO3” group, it was 95.5 % and for “AO1,” it was 93.1 %, whereas the lowest percentage was observed for “AO2” was only 58.3 %. SIMCA method confirms the possibility of easy separation of “OO” samples among different cheese samples. Additionally, Cooman’s plot, which shows the object-to-model distance for both the new objects as well as the calibration objects, was prepared (Fig. 4). Figure 4 shows the distance between the samples according to the “AO1” and “OO” model. All the “OO” are out of critical values (represents 95 % confidence interval), and there are no additional samples classified as “AO1” and any detached samples. While to the “AO1” class, both “AO3” and “AO2” samples were classified. Although there is a possibility to build the Cooman’s plot based on different models, the presented one shows good classification ability of SIMCA method. Comparing results obtained for SIMCA and LDA method (for 10 PCs), it should be affirmed that both methods had the same classification propriety for PDO-labeled cheeses (100 %). These results confirmed the effectiveness of adopted methods for identification and authenticity confirmation of PDO cheese. In the case of “AO1” samples, slightly higher classification propriety has LDA method 96.6 %, then SIMCA 93.1 %. Identical percentage of correct classification was observed for “AO3” samples (95.5 %), whereas the biggest differences were reported for “AO2” cheeses, 83.3 and 58.3 % in LDA and SIMCA analysis, respectively.
Support Vectors Machines
Support vector machine (SVM) is a binary classification tool that performs classification by constructing an optimal hyperplane, which is defined as the one that maximizes the distance between the objects and the two classes (Vapnik 1995). For the SVM analysis, 10 predictors (principal components) were used. The complete number of samples was 94, including 70 samples in a training set and 24 in a test set. Among different available function, radial basis function (RBF) was used. On the basis of SVM analysis, for the total number of samples, a 97.9 % of classification accuracy was obtained. The highest classification accuracy was observed for the test set, and it means that during the learning process, the optimal distribution of hyperplane was obtained, with the maximum separation distance. Only two samples, “AO3” and “AO2,” were misclassified, and it means that again industrially produced cheeses are problematic.
Conclusions
The main conclusion of our study is that the volatile profiles of different cheeses are specific enough and allows for the discrimination of traditional PDO Oscypek from Oscypek-like cheeses of different milk origin. In general, all applied models showed good efficiency in separation of original Oscypek from other samples. As the preprocessing of data is the same for all statistical approaches and the results are comparable, it is a matter of robustness of software package, its availability, and personal preferences which statistical method should be used.
Additionally, SPME-MS method together with chemometrics demonstrated to be a useful and effective tool in cheese discrimination and classification process. These combined methods could be irreplaceable in research on origin and authenticity of cheeses. It let to verify the honest producers, who have the certificates and declare the originality and product conformity to European Community requirements.
References
Adda J (1986) Flavour of dairy products. In: Birch GG, Lindley MD (eds) Developments in food flavours. Elsevier Applied Science, London, pp 151–172
Barron LJR, Redondo Y, Aramburu M, Perez-Elortondo FJ, Albisu M, Najera AI, de Renobables M, Fernandez-Garcia E (2005) Variations in volatile compounds and flavor in Idiazabal cheese manufactured from ewe’s milk in farmhouse and factory. J Sci Food Agric 85:1660–1671
Berezińska A, Obiedziński MW (2007) The SPME technique as a useful tool to construct aromagrams of blue-veined cheese. Food Sci Technol Qual 6:251–257
Berrueta LA, Alonso-Salces RM, Herberger K (2007) Supervised patter recognition in food analysis. J Chromatogr A 1158:196–214
Cajka T, Riddellova K, Klimankova E, Cerna M, Pudil F, Hajslova J (2010a) Traceability of olive oil based on volatiles pattern and multivariate analysis. Food Chem 121:282–289
Cajka T, Riddellova K, Tomaninova M, Hajslova J (2010b) Recognition of beer brand based on multivariate analysis of volatile fingerprint. J Chromatogr A 1217:4195–4203
Commission Regulation (EC) 2006 (http://eurlex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:C:2006:180:0094:0097:PL:PDF)
Dittmann B, Nitz G, Horner G (1998) A new chemical sensor on a mass spectrometric basis. Adv Food Sci (CTML) 20(3-4):115–121
Guerreiro JS, Barros M, Fernandes P, Pires P, Bardsley R (2013) Principal component analysis of proteolytic profiles as markers of authenticity of PDO cheeses. Food Chem 136:1526–1532
Hrbek V, Vaclavik L, Elich O, Hajslova J (2014) Authentication of milk and milk-based foods by direct analysis in real time ionization-high resolution mass spectrometry (DART-HRMS) technique: a critical assessment. Food Control 36:138–145
Jeleń HH, Ziółkowska A, Kaczmarek A (2010) Identification of botanical origin of raw spirits produced from rye, potato, and corn based on volatile compounds analysis using a SPME-MS method. J Agric Food Chem 58:12585–12591
Lv S-D, Wu Y-S, Song Y-Z, Zhou J-S, Lian M, Wang C, Liu L, Meng Q-X (2014) Multivariate analysis based on GC-MS fingerprint and volatile composition for the quality evaluation of Pu-Erh green tea. Food Anal Methods 7:1840–1847
Mafra I, Ferreira IM, Oliveira BP (2008) Food authentication by PCR-based methods. Eur Food Res Technol 227:649–665
Majcher MA, Jeleń HH (2011) Key odorants of Oscypek, a traditional polish ewe’s milk cheese. J Agric Food Chem 59:4932–4937
Majcher M, Ławrowski P, Jeleń H (2010) Comparison of original and adulterated Oscypek cheese based on volatile and sensory profiles. Acta Sci Pol Technol Aliment 9:265–275
Majcher MA, Goderska K, Pikul J, Jeleń HH (2011) Changes in volatile, sensory and microbial profiles during preparation of smoked ewe cheese. J Sci Food Agric 91:1416–1423
Mallia S, Fernandez-Garcia E, Bosset JO (2005) Comparison of purge and trap and solid phase microextraction techniques for studying the volatile aroma compounds of three European PDO hard cheeses. Int Dairy J 15:741–758
Marsili RT (1999) SPME-MS-MVA as an electronic nose for the study of off-flavors in milk. J Agric Food Chem 47:648–654
Moio L, Rillo L, Ledda A, Addeo F (1996) Odorous constituents of ovine milk in relationship to diet. J Dairy Sci 79:1322–1331
Peres C, Viallon C, Berdague JL (2001) Solid-phase microextraction-mass spectrometry: a new approach to the rapid characterization of cheeses. Anal Chem 73:1030–1036
Pillonel L, Ampuero S, Tabacchi R, Bosset JO (2003) Analytical methods for the determination of the geographic origin of Emmental cheese: volatile compounds by GC/MS-FID and electronic nose. Eur Food Res Technol 216:179–183
Stanimirova I, Daszykowski M, Walczak B (2007) Dealing with missing values and outliers in principal component analysis. Talanta 72:172–178
Stanimirova I, Ustun B, Cajka T, Riddelova K, Hajslova J, Buydens LMC, Walczak B (2010) Tracing the geographical origin of honeys based on volatile compounds profiles assessment using pattern recognition techniques. Food Chem 118:171–176
Tu D, Li H, Wu Z, Zhao B, Li Y (2014) Application of headspace solid-phase microextraction and multivariate analysis for the differentiation between edible oils and waste cooking oil. Food Anal Methods 7:1263–1270
Urbach G (1997) The flavour of milk and dairy products: II. Cheese: contribution of volatile compounds. Int J Dairy Technol 50:79–89
Urgeghe PP, Piga C, Addis M, Di Salvo R, Piredda G, Scintu MF, Wolf IV, Sanna G (2012) SPME/GC-MS characterization of the volatile fraction of an Italian PDO sheep cheese to prevalent lypolitic ripening: the case of Fiore Sardo. Food Anal Methods 5:723–730
Vandeginste BMG, Massart DL, Buydens LMC, de Jong S, Lewi PJ, Smeyers-Verbeke J (1998) Handbook of chemometrics and qualimetrics. Part B. Elsevier, Amsterdam
Vapnik V (1995) The nature of statistical learning theory. Springer, New York
Wold S (1976) Pattern recognition by means of disjoint principal component models. Pattern Recogn 8:127–139
Acknowledgments
Polish State Committee for Scientific Research financed this research project (No. N N312 1571134).
Conflict of Interest
Małgorzata Majcher declares that she has no conflict of interest. Anna Kaczmarek declares that she has no conflict of interest. Dorota Klensporf-Pawlik declares that she has no conflict of interest. Jan Pikul declares that he has no conflict of interest. Henryk Jeleń declares that he has no conflict of interest. We, the authors, declare that this article does not contain any studies with human or animal subjects.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Majcher, M.A., Kaczmarek, A., Klensporf-Pawlik, D. et al. SPME-MS-Based Electronic Nose as a Tool for Determination of Authenticity of PDO Cheese, Oscypek. Food Anal. Methods 8, 2211–2217 (2015). https://doi.org/10.1007/s12161-015-0114-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12161-015-0114-x