Abstract
Alpha1-antitrypsin is a 45-kDa (394-residue) serine protease inhibitor synthesized by hepatocytes, which is released into the circulatory system and protects the lung from the actions of neutrophil elastase via a conformational transition within a dynamic inhibitory mechanism. Relatively common point mutations subvert this transition, causing polymerisation of α1-antitrypsin and deficiency of the circulating protein, predisposing carriers to severe lung and liver disease. We have assigned the backbone resonances of α1-antitrypsin using multidimensional heteronuclear NMR spectroscopy. These assignments provide the starting point for a detailed solution state characterization of the structural properties of this highly dynamic protein via NMR methods.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Biological context
Alpha1-antitrypsin is a glycoprotein of the serpin (serine protease inhibitor) protein superfamily (Silverman et al. 2001). It is synthesized within hepatocytes and secreted into the circulatory system, where it is the most abundant human plasma proteinase (Gooptu and Lomas 2009; Travis and Salvesen 1983). The primary function of α1-antitrypsin is the inhibition of neutrophil elastase. Naturally occurring point mutations within the protein such as the Z (Glu342Lys) variant cause it to undergo conformational rearrangement and self-associate into polymer chains (Lomas et al. 1992). The accumulation of α1-antitrypsin polymers within the endoplasmic reticulum (ER) is associated with cellular toxicity and development of liver diseases such as cirrhosis and hepatocellular carcinoma (Eriksson et al. 1986; Perlmutter 2002). Polymerisation abolishes functional activity of α1-antitrypsin and so allows dysregulated elastase activity within the lung, causing emphysema (Elliott et al. 1998). Moreover, extracellular α1-antitrypsin polymers are found within the lung in patients with α1-antitrypsin deficiency related emphysema, and are likely to have pro-inflammatory effects. The combination of loss- and gain-of-function consequences of α1-antitrypsin polymerisation mean there is a need for a strategy that prevents polymerisation and maintains enzyme inhibitory activity (Gooptu et al. 2009). While some small molecules have shown polymerisation blocking effects in vitro, so far they have all abolished the functional activity of α1-antitrypsin (Mahadeva et al. 2002; Mallya et al. 2007). Moreover, there is a lack of structural data defining interactions of the protein with promising lead compounds, preventing their rational modification. Solution NMR is well-suited to provide a residue-specific characterisation of this conformationally heterogeneous protein and to probe its interactions with putative polymerisation blockers. We report here the 1H, 15N and 13C backbone assignments of 45 kDa α1-antitrypsin as a crucial step not only for detailed characterization of this highly dynamic molecule but also towards the application of drug discovery techniques to α1-antitrypsin deficiency.
Methods and experiments
cDNA coding for 6xHis-tagged α1-antitrypsin within a pQE31 plasmid was transformed into BL21-Gold (DE3) E. coli (Stratagene). Cells were grown in M9 minimal medium (H2O or D2O) at 37°C with 1 g/l of 15NH4Cl (Spectra Stable Isotopes) and 2 g/l of either glucose or 13C-glucose (Sigma-Aldrich) as the sole nitrogen and carbon sources, respectively. Following induction of protein expression at OD600 = 0.6 using isopropyl β-D-1-thiogalactopyranoside (IPTG), cells were incubated at 30°C for 8 h. The cells were collected by centrifugation, resuspended in 10 mM sodium phosphate buffer, pH 8.0, 0.5 M NaCl, 20 mM imidazole, and lysed using a cell disruptor. The supernatant of the cell lysate was loaded onto a HisTrap Crude FF column (5 ml; GE Healthcare) and after washing to baseline with the same buffer, the bound protein was eluted with an imidazole gradient (20 mM – 200 mM). The fractions containing α1-antitrypsin were collected, dialyzed 10 mM Tris, pH 8.0, 1 mM EDTA, and 1 mM β-mercaptoethanol and loaded onto a HiTrap-Q Sepharose column (5 ml; GE Healthcare). Bound protein was then eluted with a 0–0.5 M NaCl gradient. The fractions containing α1-antitrypsin were pooled, dialysed against 25 mM Na2HPO4, 50 mM NaCl and 1 mM EDTA at pH 8.0. For complete back exchange of buried backbone amides in 2H/15N/13C labeled α1-antitrypsin, the protein (1 mg/ml) was unfolded in storage buffer containing 8 M urea for 1 h at room temperature. It was subsequently refolded by dropwise addition to 5 L storage buffer with continuous stirring at 4°C. Precipitated material was removed by centrifugation, and the conformational integrity of the refolded protein was confirmed by non-denaturing and SDS-PAGE and inspection of its 15N TROSY-HSQC spectrum.
NMR spectra were collected from α1-antitrypsin at a typical concentration of 225 μM in 25 mM Na2HPO4, 50 mM NaCl, 1 mM EDTA and 0.001% 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS) at pH 8.0 or pH 7.0 (10% D2O/90% H2O). Spectra from a sample prior to refolding at pH 8.0 were recorded on a Bruker Avance III 700 MHz spectrometer equipped with an HCN cryoprobe. TROSY (Pershuvin et al. 1997) versions of HNCO, HN(CA)CO, HNCA, HN(CO)CA, HNCACB and HN(CO)CACB spectra (Eletsk et al. 2001; Salzmann et al. 1998, 1999) were acquired with constant-time chemical shift evolution in the 15N dimension and 2H decoupling (1 kHz WALTZ-16) (Shaka et al. 1983) while magnetisation was transverse on CA/CB. Spectra from refolded samples at pH 8.0 and pH 7.0 were acquired on Varian Inova 600, 800 and 900 MHz spectrometers, all equipped with HCN cold-probes. TROSY-HNCA (Yang and Kay 1999) spectra were acquired with 2H decoupling as above and constant-time chemical shift evolution in the 13C indirect dimension. All spectra were processed using NMRPipe (Delaglio et al. 1995) and analysed using the CCPN software suite (Fogh et al. 2002).
Assignments and data deposition
The sequential backbone resonance assignments of α1-antitrypsin at pH 8.0 and pH 7.0 have been deposited in the Biological Magnetic Resonance Data Bank (http://www.bmrb.wisc.edu) under the accession number 17804. The 15N-HSQC spectrum of 45 kDa α1-antitrypsin shows excellent cross-peak dispersion (Fig. 1). With 394 residues, α1-antitrypsin is to date one of the largest proteins for which the backbone has been assigned to near completion (Supp Fig. 1). The high molecular weight rendered the backbone assignment a challenging task and this was therefore carried out in successive stages. Initially (stage 1), native 2H/15N/13C-labelled antitrypsin was used, followed by the use of refolded (see Methods) 2H/15N/13C antitrypsin samples at pH 8.0 (stage 2) and pH 7.0 (stage 3). The majority of the secondary-structure elements were assigned in stage 1, except for resonances corresponding to the core β-sheet B and also the loop regions (Supp Fig. 2a), which were assigned during stages 2 (Supp Figs. 2b and 3a) and 3 (Supp Figs. 2c and 3b), respectively. Aside from the additional expected cross-peaks, the 15N-TROSY-HSQC spectra before and after refolding were identical.
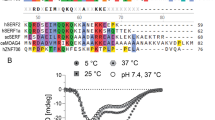
82.8 and 79.3% of backbone amide resonances were assigned at pH 8.0 and pH 7.0, respectively. 84.9, 88.3 and 75.1% of the CO, CA and CB resonances, respectively, were assigned at pH 8.0. At pH 7.0, 82.5% of the CA resonances were assigned. The secondary structure of a1-antitrypsin was predicted from the assigned chemical shifts at pH 8.0 using DANGLE (Cheung et al. 2010). The measured CA and CB chemical shifts were first corrected for deuterium isotope shifts (Venters et al. 1996; Gardner et al. 1997). The predicted secondary structure is broadly consistent with that observed crystallographically (Elliott et al. 2000; Fig. 2).
Predicted secondary-structure of α1-antitrypsin. The secondary structure of the crystal structure (PDB ID 1QLP) is shown schematically and compared with the secondary structure predicted from the assigned chemical shift (CA, CB, CO and NH) using the program DANGLE (where C = random coil, H = α-helix, E = β-strand, N = no prediction). No predictions are shown for residues that are not fully assigned
References
Cheung M-S, Maguire ML, Stevens TJ, Broadhurst RW (2010) DANGLE: a Bayesian inferential method for predicting protein backbone dihedral angles and secondary structure. J Magn Reson 202:223–233
Delaglio F, Grzesiek S, Vuister G, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293
Eletsk A, Kienhofer A, Pervushin K (2001) TROSY NMR with partially deuterated proteins. J Biomol NMR 20:177–180
Elliott PR, Bilton D, Lomas DA (1998) Lung polymers in Z α1-antitrypsin related emphysema. Am J Respir Cell Mol Biol 18:670–674
Elliott PR, Pei XY, Dafforn TR, Lomas DA (2000) Topography of a 2.0Å structure of α1-antitrypsin reveals targets for rational drug design to prevent conformational disease. Protein Sci 9:1274–1281
Eriksson S, Carlson J, Velez R (1986) Risk of cirrhosis and primary liver cancer in alpha1-antitrypsin deficiency. N Engl J Med 18:670–674
Fogh R, Ionides J, Ulrich E, Boucher W, Vranken W, Linge JP, Habeck M, Rieping W, Bhat TN, Westbrook J, Henrick K, Gilliland G, Berman H, Thornton J, Nilges M, Markley J, Laue E (2002) The CCPN project: an interim report on a data model for the NMR community. Nat Struct Biol 9:416–418
Gardner KH, Rosen MK, Kay LE (1997) Global folds of highly deuterated, methyl-protonated proteins by multidimensional NMR. Biochemistry 36:1389–1401
Gooptu B, Lomas DA (2009) Conformational pathology of the serpins: themes, variations, and therapeutic strategies. Annu Rev Biochem 78:147–176
Gooptu B, Ekeowa UI, Lomas DA (2009) Mechanisms of emphysema in α1-antitrypsin deficiency: molecular and cellular insights. Eur Respir J 34:475–488
Lomas DA, Evans DL, Finch JT, Carrell RW (1992) The mechanism of Z α1-antitrypsin accumulation in the liver. Nature 357:605–607
Mahadeva R, Dafforn TR, Carrell RW, Lomas DA (2002) Six-mer peptide selectively anneals to a pathogenic serpin conformation and blocks polymerisation: implications for the prevention of Z α1-antitrypsin related cirrhosis. J Biol Chem 277:6771–6774
Mallya M, Phillips RL, Saldanha SA, Gooptu B, Leigh Brown SC, Termine DJ, Shirvani AM, Wu Y, Sifers RN, Abagyan R, Lomas DA (2007) Small molecules block the polymerization of Z alpha1-antitrypsin and increase the clearance of intracellular aggregates. J Med Chem 50:5357–5363
Perlmutter DH (2002) Liver injury in α1-antitrypsin deficiency: an aggregated protein induces mitochondrial injury. J Clin Investig 110:1579–1583
Pershuvin K, Riek R, Wider G, Wuthrich K (1997) Attenuated T2 relaxation by mutual cancellation of dipole–dipole coupling and chemical shift anisotropy indicates and avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci USA 94:12366–12371
Salzmann M, Pershuvin K, Wider G, Senn H, Wuthrich K (1998) TROSY in triple-resonance experiments: new perspectives for sequential NMR assignment of large proteins. Proc Natl Acad Sci USA 95:13585–13590
Salzmann M, Wider G, Pervushin K, Senn H, Wüthrich K (1999) TROSY-type triple-resonance experiments for sequential NMR assignments of large proteins. J Am Chem Soc 121:844–848
Shaka AJ, Keeler J, Frenkiel T, Freeman R (1983) An improved sequence for broadband decoupling: WALTZ-16. J Magn Reson 52:335–338
Silverman GA, Bird PI, Carrell RW, Church FC, Coughlin PB, Gettins PGW, Irving JA, Lomas DA, Luke CJ, Moyer RW, Pemberton PA, Remold-O’Donnell E, Salvesen GS, Travis J, Whisstockc JC (2001) The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. J Biol Chem 276:33293–33296
Travis J, Salvesen GS (1983) Human plasma proteinase inhibitors. Ann Rev Biochem 52:655–709
Venters RA, Farmer BT II, Fierke CA, Spicer LD (1996) Characterizing the use of perdeuteration in NMR studies of large proteins: 13C, 15N and 1H assignments of human carbonic anhydrase II. J Mol Biol 264:1101–1116
Yang D, Kay LE (1999) Improved 1HN-detected triple resonance TROSY-based experiments. J Biomol NMR 13:3–10
Acknowledgments
We are grateful for the use of the Biological NMR Centre at UCL, the Henry Wellcome Building Biomolecular NMR Facility, University of Birmingham and the MRC Biomedical NMR Centre (Mill Hill). This work was supported by funding from the Wellcome Trust, the Medical Research Council (UK), the Human Frontiers Science Programme, the Biotechnology and Biological Sciences Research Council, Papworth Hospital NHS Trust, and the Birkbeck College Faculty Research Fund.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding authors
Additional information
M. P. Nyon, J. Kirkpatrick contributed equally.
Electronic supplementary material
Below is the link to the electronic supplementary material.
12104_2011_9345_MOESM1_ESM.tif
Histogram of the sizes of entries containing 15N assignments in the BioMagResBank (BMRB) as of July 2011. The histogram is plotted with a bin width of ten 15N resonances. The arrow locates the alpha1-antitrypsin assignment. The bar with asterisk represents entries with more than 350 assigned 15N resonances (9 entries). (TIFF 442 kb)
12104_2011_9345_MOESM2_ESM.tif
Residues with backbone 1H-15N assignments are highlighted in blue, not assigned are in red and prolines are in green: (a) stage 1 at pH 8.0 (b) stage 2 at pH 8.0 with refolded sample (c) stage 3 at pH 7.0. (TIFF 5217 kb)
12104_2011_9345_MOESM3_ESM.tif
The labelled cross peaks are the additional assignments (a) at pH 8.0 obtained after refolding the protein (b) after lowering the pH from 8.0 to 7.0. (TIFF 2903 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Nyon, M.P., Kirkpatrick, J., Cabrita, L.D. et al. 1H, 15N and 13C backbone resonance assignments of the archetypal serpin α1-antitrypsin. Biomol NMR Assign 6, 153–156 (2012). https://doi.org/10.1007/s12104-011-9345-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-011-9345-y