Abstract
Purpose
Esophageal squamous cell carcinoma (ESCC) is one of the most important causes of mortality in the developing world. Although hereditary forms arise from germ-line mutations in TP53, Rb, and the mismatch repair genes, many familial cases present with an unknown inherited cause. The new theory of rare, high-penetrance mutations in less known genes is a likely explanation for the underlying predisposition in some of these familial cases.
Methods
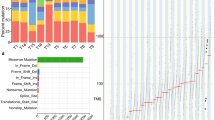
Exome sequencing was performed in 9 patients with esophageal squamous cancer from 9 families with strong disease aggregation without mutations in known hereditary esophageal cancer genes. Data analysis was limited to only really rare variants (0–0.01%), producing a putative loss of function and located in genes with a role compatible with carcinogenesis.
Results
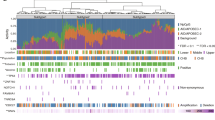
Twenty-two final candidate variants were selected and validated by Sanger sequencing. Correct family segregation and somatic studies were used to categorize the most interesting variants in CDK11A, ARID1A, JMJD6, MAML3, CDKN2AIP, and PHLDA1.
Conclusion
Together, we identified new potential esophageal squamous cancer predisposition variants in genes which may have a role in cancer and are involved in chromatin remodeling and cell-cycle pathway, which could increase the risk of ESCC.


Similar content being viewed by others
Availability of data and materials
Imaging data could be provided upon request.
Abbreviations
- ESCC:
-
Esophageal squamous cell carcinoma
- WES:
-
Whole-exome sequencing
- MAML3:
-
Mastermind-like 3
References
Simard EP, et al. Cancers with increasing incidence trends in the United States: 1999 through 2008. CA A Cancer J Clin. 2012;62(2):118–28.
Wen X-D, et al. Earlier onset and multiple primaries in familial as opposed to sporadic esophageal cancer. World. 2014;2:005.
Stoner GD, Gupta A. Etiology and chemoprevention of esophageal squamous cell carcinoma. Carcinogenesis. 2001;22(11):1737–46.
Mir MM, et al. p53 mutation profile of squamous cell carcinomas of the esophagus in Kashmir (India): a high-incidence area. Int J Cancer. 2005;116(1):62–8.
Su H, et al. Gene expression analysis of esophageal squamous cell carcinoma reveals consistent molecular profiles related to a family history of upper gastrointestinal cancer. Can Res. 2003;63(14):3872–6.
Albertson DG. Gene amplification in cancer. Trends Genet. 2006;22(8):447–55.
Pollack JR, et al. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet. 1999;23(1):41.
Huang FL, Yu SJ. Esophageal cancer: risk factors, genetic association, and treatment. Asian J Surg. 2018;41(3):210–5.
Marco-Sola S, et al. The GEM mapper: fast, accurate and versatile alignment by filtration. Nat Methods. 2012;9(12):1185.
Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–9.
Fuentes Fajardo KV, et al. Detecting false-positive signals in exome sequencing. Hum Mutat. 2012;33(4):609–13.
Esteban-Jurado C, et al. Whole-exome sequencing identifies rare pathogenic variants in new predisposition genes for familial colorectal cancer. Genet Med. 2015;17(2):131.
Liu T-H, et al. Identification and characterization of the BmCyclin L1-BmCDK11A/B complex in relation to cell cycle regulation. Cell Cycle. 2017;16(9):861–8.
Shi J, Nelson MA. The cyclin-dependent kinase 11 interacts with NOT2. Biochem Biophys Res Commun. 2005;334(4):1310–6.
Lahti JM, et al. Alterations in the PITSLRE protein kinase gene complex on chromosome 1p36 in childhood neuroblastoma. Nat Genet. 1994;7(3):370.
Bonatto N, et al. PHLDA1 (pleckstrin homology-like domain, family A, member 1) knockdown promotes migration and invasion of MCF10A breast epithelial cells. Cell Adhes Migr. 2018;12(1):37–46.
Cheung CT, et al. CARF: an emerging regulator of p53 tumor suppressor and senescence pathway. Mech Ageing Dev. 2009;130(1–2):18–23.
Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11(7):481.
Xiaomei W, et al. Two related ARID family proteins are alternative subunits of human SWI/SNF complexes. Biochem J. 2004;383(2):319–25.
Weissman B, Knudsen KE. Hijacking the chromatin remodeling machinery: impact of SWI/SNF perturbations in cancer. Can Res. 2009;69(21):8223–300.
Oyama T, et al. Mastermind-like 1 (MamL1) and mastermind-like 3 (MamL3) are essential for Notch signaling in vivo. Development. 2011;138(23):5235–46.
Wu L, et al. Identification of a family of mastermind-like transcriptional coactivators for mammalian notch receptors. Mol Cell Biol. 2002;22(21):7688–700.
Li J, et al. MiR-2392 suppresses metastasis and epithelial–mesenchymal transition by targeting MAML3 and WHSC1 in gastric cancer. FASEB J. 2017;31(9):3774–866.
Mantri M, et al. Crystal structure of the 2-oxoglutarate-and Fe (II)-dependent lysyl hydroxylase JMJD6. J Mol Biol. 2010;401(2):211–22.
Unoki M, et al. Lysyl 5-hydroxylation, a novel histone modification, by Jumonji domain containing 6 (JMJD6). J Biol Chem. 2013;288(9):6053–62.
Wang F, et al. JMJD6 promotes colon carcinogenesis through negative regulation of p53 by hydroxylation. PLoS Biol. 2014;12(3):e1001819.
Hong X, et al. Interaction of JMJD6 with single-stranded RNA. Proc Natl Acad Sci. 2010;107(33):14568–72.
Clissold PM, Ponting CP. JmjC: cupin metalloenzyme-like domains in jumonji, hairless and phospholipase A2β. Trends Biochem Sci. 2001;26(1):7–9.
Webby CJ, et al. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science. 2009;325(5936):90–3.
Jones S, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330(6001):228–31.
Wiegand KC, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363(16):1532–43.
Katoh M. Dysregulation of stem cell signaling network due to germline mutation, SNP, Helicobacter pylori infection, epigenetic change, and genetic alteration in gastric cancer. Cancer Biol Ther. 2007;6(6):832–9.
Chang B, et al. JMJD6 is a histone arginine demethylase. Science. 2007;318(5849):444–7.
Acknowledgements
We thank our colleagues in Avicenna research center for sharing information from data bank.
Funding
The author(s) received no financial support for the research, authorship, and/or publication of this article.
Author information
Authors and Affiliations
Contributions
FFG and MRA design the experiment, analysis the data, and FFG and TED draft the paper for the work. MRA and TED help to revise the paper critically for important intellectual content. MRA did the financial support, review, and final approval of the paper to be published. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethics approval
This study was approved by the Mashhad University of Medical Science (MUMS).
Informed consent
Patient consent was obtained prior to the initiation of the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Golyan, F.F., Druley, T.E. & Abbaszadegan, M.R. Whole-exome sequencing of familial esophageal squamous cell carcinoma identified rare pathogenic variants in new predisposition genes. Clin Transl Oncol 22, 681–693 (2020). https://doi.org/10.1007/s12094-019-02174-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-019-02174-z