Abstract
Connective tissue growth factor (CTGF, CCN2) is overexpressed in pancreatic cancer. We mapped the minimal CCN2 promoter active in PANC-1 cells, a human pancreatic cancer cell line. Within this region, Sp1, BCE-1 and Ets elements were important for the activity of the CCN2 promoter. Constitutive hyperactivated ras is a hallmark of cancers, including that of the pancreas. Treatment of PANC-1 cells with the MEK inhibitor U0126 or the Sp1 inhibitor mithramycin reduced CCN2 mRNA and promoter activity. Mutation of the BCE-1, but not Sp1 or Ets, site abolished the responsiveness of the CCN2 promoter to U0126. Overexpressing constitutively active MEK1 or ras activated CCN2 promoter activity. Thus CCN2 is likely to act downstream of ras in PANC-1 cells. CCN2 is overexpressed in cancer cells. Activated ras/MEK/ERK is a hallmark of cancer, and we have shown that the elevated CCN2 expression in pancreatic cancer cells is dependent on this pathway.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pancreatic cancer is currently the fourth leading cause of cancer deaths in North America, with a median survival rate of 6.4 months (Aikawa et al. 2006). Previous studies have shown that pancreatic tumour cells have 59 times the normal amount of connective tissue growth factor (CTGF/CCN2; Wenger et al. 1999). CCN2 transcript levels are proportional to the severity of pancreatic cancer (Wenger et al. 1999; Hartel et al. 2004). CCN2 is a protein found in the extracellular matrix (ECM) and functions as a modifier of adhesive signalling in response to ECM and cytokines (Chen et al. 2004; Shi-wen et al. 2006). CCN2 acts through integrins and heparin-sulphate containing proteoglycans influencing cell growth, apoptosis, migration and ECM remodelling and deposition (Leask and Abraham 2006). CCN2 promotes angiogenesis in vivo and in vitro (Babic et al. 1999). Loss of CCN2 expression in embryonic mesenchymal cells results in diminished cell migration and adhesion, and reduction in pro-angiogenic and pro-fibrotic gene expression (Chen et al. 2004; Kennedy et al. 2007). Previous studies have shown that application of an antibody specific to CCN2 resulted in decreased tumour volume, metastasis and angiogenesis (Aikawa et al. 2006).
CCN2 expression is controlled primarily at the transcriptional level; therefore elements in its promoter are likely to contribute to its overexpression in pancreatic cancer (Blom et al. 2001). However, it is unknown which cis-acting elements are responsible. Previously, we have analysed the regions in the CCN2 promoter required for its induction in response to TGFβ and endothelin-1 in normal fibroblasts and mesangial cells, and its constitutively elevated expression in fibrotic scleroderma fibroblasts (Abraham et al. 2000; Holmes et al. 2001, 2003; Leask et al. 2003; van Beek et al. 2006). CCN2 induction in response to TGFβ in fibroblasts and mesangial cells requires Smad 3 and Ets-1 and also requires ras/MEK/ERK (Holmes et al. 2001; Stratton et al. 2002; Chen et al. 2002; Leask et al. 2003; van Beek et al. 2006). In addition, a basal control element (BCE-1) and Sp1 contribute to basal activity of the CCN2 minimal promoter and the elevated CCN2 promoter activity in scleroderma (Holmes et al. 2001, ).
Although CCN2 appears important in pancreatic cancer progression, a mechanism for its overexpression in this disease has yet to be proposed. Thus, we identified the minimal CCN2 promoter active in PANC-1 pancreatic cancer cells, and probed the cis-acting sequences and signalling cascades required for its activity.
Materials and methods
Materials
PANC-1 cells were obtained from Christopher Pin (University of Western Ontario). High glucose Dulbecco’s modified Eagle’s medium (DMEM), trypsin-EDTA, fetal bovine serum (FBS), antibiotic/antimycotic solution were all obtained from Invitrogen (Burlington, Ontario).
Cell cultures, transfections and DNA constructs
PANC-1 cells were o cultured in high glucose DMEM (Invitrogen, Burlington, Ontario). All media was supplemented with 10% FBS and 1% antibiotic/antimycotic solution. For media assays, PANC-1 cells were plated at a density of 25,000 cells/well in a 24 well plate. Cells were allowed to grow for 24 h at 37°C. Cells were then transfected with Fugene (Roche, Indianapolis, IN, USA) at a ratio of 3:25 μl DMEM per well. Cells were transfected with plasmids containing a CCN2 promoter fused to a secreted alkaline phosphatase (SEAP) reporter gene. Cells were transfected with 2 μg of CTGF-SEAP constructs that were contained either −244, −166 or −86 to +17 region of the CCN2 promoter, or had a point mutation to either the Smad, Ets, BCE-1 or Sp1 site (Abraham et al. 2000; Holmes et al. 2001, 2003; Leask et al. 2003). When indicated, cells were transfected with empty expression vector, or expression vectors encoding constitutively activated ras or MEK1 (Upstate). To control for transfection efficiency cells were transfected with 0.4 μg of a cytomegalovirus (CMV) promoter – β-galactosidase (β-gal) reporter gene (Clontech, Palo Alto, CA, USA) construct. Cells were allowed to grow for 48 h in 37°C. Inhibitors (when used) were added 24 h after transfection. Promoter assays were performed with a Phospha-Light kit (Applied Biosystems, Foster City, CA, USA) according to manufacturer’s protocol and SEAP reporter expression was adjusted for differences in β-galactosidase expression as determined by a Galacto-star kit (Applied Biosystems) according to manufacturer’s protocol. Data was expressed as average values +/− standard deviation of at least three replicates and at least two independent trials. Measurement of SEAP levels were obtained from an LMax II 384 luminometer (Molecular Devices, Sunnyvale CA, USA) and SoftMax Pro 4.7.1 (Molecular Devices, Sunnyvale CA, USA). Samples were run in quadruplicate, and levels were measured in relative light units and were standardized to control values from â-gal. Statistical tests were done using one-way ANOVA and Tukey’s post hoc test on GraphPad.
Real time RT-PCR
PANC-1 cells were grown in high glucose DMEM. All media was supplemented with 10% FBS and 1% antibiotic/antimycotic. Cells were plated onto a six well plate at a density of 60,000 cells/well and were allowed to grow for 24 h at 37°C. Cells were then treated with focal adhesion kinase/src inhibitor (FAK/SRC) PP2, p38 MAP Kinase inhibitor SB203580, MEK/ERK inhibitor U0126, phosphatidyl-inositol triphosphate (PIP3) inhibitor LY294002, protein kinase C (PKC) inhibitor bisindolmaleimide I (all Calbiochem, La Jolla, CA, USA), or Sp1 inhibitor Mithramycin (Sigma, St Louis, MO, USA) at a concentration of 10 μM, except mithramycin which was used at 100 nM and SB203580 which was used at 30 μM. RNA was harvested using the Qiashredder and RNeasy kit (Qiagen) and used for real-time RT-PCR. Twenty-five nanograms of RNA was reverse transcribed and amplified using TaqMan Assays on Demand (Applied Biosystems) in a 15 μl reaction containing primers for TaqMan Human CTGF (Applied Biosystems) and 6-carboxyfluroscein labelled TaqMan MGB probe. Reverse transcriptase qPCR Mastermix (Eurogenetics) was added to samples and the ABI Prism 7900 HT sequence detector (Perkin-Elmer-Cetus, Vaudreuil, QC) was used according to manufacturer’s instructions to detect amplified sequences. Samples were run in triplicate, transcripts were measured in picograms and expression values were standardized to control values from GAPDH primers. Statistical analysis was done using one way ANOVA and Tukey’s post hoc test on GraphPad.
Results
Ets, BCE-1 and Sp1, but not Smad, sites contribute to CCN2 minimal promoter activity in PANC-1 cells
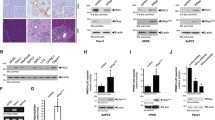
To determine which promoter elements contributed to CCN2 overexpression in PANC-1 cells, PANC-1 cells were transfected with CCN2 promoter-SEAP reporter constructs containing regions spanning between −244, −166 or −86 to +17. The −244 to +17 promoter fragment contains the previously identified CCN2 minimal promoter region (Leask et al. 2003) and Smad, BCE-1, Ets-1 and Sp1 sites (Fig. 1a). Conversely, the −166 promoter contained only BCE-1, Ets-1 and Sp1 and the −86 promoter contained only the Sp1 region (Leask et al. 2003). Although all three promoter fragments possessed abundant activity, only deleting −166 to −86 had a statistically significant impact on basal CCN2 promoter activity in PANC-1 cells (Fig. 1).
Identification of the CCN2 minimal promoter in PANC-1 Cells. a Diagram of the CCN2 minimal promoter, including known functional transcription factor elements. The bases mutated in each construct are shown below. Wt wild-type sequence, mut mutated sequence. b PANC-1 cells were co-transfected for 48 h with CCN2 promoter-SEAP reporter test constructs and a CMV promoter-β-Galactosidase construct. Plasmids had regions of the CCN2 promoter starting at −244, −166 or −86 to +17. Values were expressed in relative light units and normalized with β-galactosidase. Values are mean of four replicates, +/− standard deviation from three separate experiments. Asterisk indicates significantly different from −244/+17 construct, P < 0.01
To further investigate which specific promoter elements were involved in CTGF overexpression, PANC-1 cells were transfected with CCN2 promoter-SEAP reporter constructs containing a wild-type promoter sequence, or point mutations to either the Smad, BCE-1, Ets-1 or Sp1 sites. Consistent with our deletion analysis showing removal of the Smad element had no significant impact on activity of the CCN2 minimal promoter, point mutation to the Smad site resulted in no significant decrease in CCN2 promoter activity; indeed, mutating the Smad element seemed to increase basal CCN2 promoter activity in PANC-1 cells. Conversely, mutating BCE-1, Ets, or Sp1 sites resulted in significant decreases in promoter activity (Fig. 2). Moreover, treatment of PANC-1 cells with the Sp1 inhibitor mithramycin reduced expression of CCN2 mRNA and protein (Fig. 3a, b) and activity of the CCN2 minimal promoter (Fig. 4). These results suggest that the BCE-1, Ets and Sp1, but not Smad element, are implicated in CCN2 overexpression in PANC-1 cells.
Ets, BCE-1 and Sp1, but not Smad, sites contribute to CTGF promoter activity in PANC-1 cells. PANC-1 cells were co-transfected for 48 h with CCN2 promoter-SEAP reporter test constructs and a CMV promoter-β-galactosidase construct. Plasmids contained either the wild-type CCN2 promoter or otherwise identical promoters with mutations in Smad, Ets, BCE-1 or Sp1 sites and expression values were measured in relative light units. Expression values were normalized with β-galactosidase. The experiment was repeated performed twice. Values were expressed in relative light units and normalized with β-Galactosidase. Values are mean of four replicates, +/− standard deviation from three separate experiments. *significantly different from −244/+17 construct, P < 0.05. Note that mutation of Ets, BCE-1 and Sp1 sites decreased CCN2 promoter activity, whereas mutation of the Smad element increased CCN2 promoter activity
Inhibiting MEK and Sp1 potently decreased CCN2 mRNA and protein expression. a Cells were incubated for 24 h with inhibitors of signal transduction cascades. RNA was harvested and subjected to real time RT-PCR and was normalised with GAPDH. The experiment was performed twice. Columns, mean of separate experiments performed in triplicate; bars, SE (asterisk indicates significantly reduced CCN2 mRNA due to inhibitor, P < 0.001). Inhibitors used were: 10 μM bisindolmaleimide I (PKC), 10 μM LY294002 (LY), 30 μM SB203580 (SB), 10 μM PP2, 10 μM U0126 or 100 nM mithramycin. b Cells were incubated for 24 h with inhibitors of signal transduction cascades. Protein was harvested and subjected to Western blot analyses with anti-CCN2 and anti-β-actin antibodies
MEK and Sp1 significantly contribute to CTGF promoter activity. a PANC-1 cells were transfected with a CCN2 full length promoter-SEAP construct and a CMV promoter-β-galactosidase construct. Twenty-four hours after treatment with constructs, the cells were treated for 24 h with mithramycin or U0126 inhibitors. Experiments were performed twice. Expression values were normalised with β-galactosidase. Columns: means of four replicates; bars, SE, (asterisk indicates significantly reduced CTGF expression due to inhibitor, P < 0.01, double asterisks indicate significantly reduced CTGF expression due to inhibitor, P < 0.001). b PANC-1 cells were transfected with a CCN2 full length promoter-SEAP construct and a CMV promoter-β-galactosidase construct. Cells were co-transfected with empty expression vector (EMPTY), or expression vector encoding constitutively active ras (caras) or constitutively active MEK1 (caMEK1). Cells were assayed 48 h after transfection. Experiments were performed twice. Values were expressed in relative light units and normalized with β-galactosidase. Values are mean of four replicates, +/− standard deviation from three separate experiments. Asterisk indicates significantly different from −244/+17 construct, P < 0.01
Inhibiting MEK potently decreases CCN2 mRNA expression in PANC-1 cells
In order to determine which signaling pathways were most important in CCN2 overexpression in pancreatic cancer, PANC-1 cells were treated with inhibitors to p38 (SB 203580), MEK/ERK (U0126), FAK/SRC (PP2), PI3 kinase (LY 294002) or PKC (bisindolmaleimide I) at concentrations previously demonstrated to selectively block action of the respective targets. Although real-time RT-PCR analysis determined that while inhibition of p38, FAK/SRC, PI3 kinase and PKC resulted in modest (∼50–60%) reductions in CCN2 mRNA, inhibition of MEK resulted in a potent (∼90%) reduction of CCN2 mRNA expression (Fig. 3).
Based on these data showing that inhibition of MEK strongly reduced CCN2 mRNA in PANC-1 cells, and previous observations that activated ras/MEK/ERK is a key, characteristic feature of cancer cells (Yamamoto et al. 1999), we further probed the contribution of ras/MEK/ERK in regulation of CCN2. First, U0126 reduced CCN2 protein expression in PANC-1 cells (Fig. 3). Second, PANC-1 cells were transfected with CCN2 promoter-SEAP reporter constructs, and then treated with U0126. Treatment with U0126 resulted in a significant decrease in CCN2 promoter activity as compared to controls (Fig. 4a). Moreover, overexpression of either activated ras or activated MEK1 increased CCN2 promoter activity (Fig. 4b).
MEK Acts through BCE-1
As MEK appeared to be important in the activity of the CCN2 promoter in PANC-1 cells, we wanted to further investigate the mechanism by which MEK was influencing CCN2 expression. In order to determine whether MEK was acting through the BCE-1, Ets or Sp1 element, cells were transfected with CCN2 promoter-SEAP reporter constructs containing mutations in these sites. Cells in each treatment group were then further split into two separate groups: a control group and a group that was incubated with the MEK inhibitor for 24 h. Only mutation of the BCE-1 site abolished the sensitivity of the CCN2 promoter to MEK inhibition (Fig. 5). Collectively, these data suggest that ras/MEK/ERK induces the CCN2 promoter in PANC-1 cells through the BCE-1 element.
MEK acts through BCE-1 in PANC-1 cells. Cells were incubated for 48 hours with a CCN2 promoter-SEAP reporter construct that was either full length or had mutations to the BCE-1, Ets or Sp1 region, as well as with a CMV-β-galactosidase construct. Half were then incubated for 24 h with 10 μmol ml−1 media−1 of U0126. Experiments were performed twice, with four replicates per trial. Results were essentially identical in each experiment. Expression values were measured in relative light units and are displayed as a percentage of the control untreated wild-type values. (asterisk indicates significantly reduced activity of the CCN2 promoter as compared to U0126 treatment, P < 0.01). Standard deviation values were all below 10%
Discussion
Previous studies have found that CCN2 is abundant in pancreatic cancer, and that this overexpression may be related to the cancer progression (Aikawa et al. 2006; Dornhofer et al. 2006). However, until this report there was no proposed mechanism for CCN2 overexpression in pancreatic cancer. As there is currently no sufficient treatment for pancreatic cancer, obtaining a greater understanding of the mechanisms underlying pancreatic cancer progression, such as the mechanism underlying the overexpression of CCN2, is crucial in order to identify novel potential drug targets.
It has been shown previously in fibroblasts and mesenchymal cells that CCN2 is induced by TGF-β acting through the Smad 3/4 pathway and an Ets element (Leask et al. 2003). However, in this report we showed that the increased basal activity of the CCN2 promoter in PANC-1 was not diminished by mutation or deletion of the Smad element. Conversely, mutation of the Ets, Sp1 or BCE-1 elements reduced the activity of the CCN2 promoter in PANC-1 cells. In addition, the Sp1 inhibitor mithramycin reduced CCN2 mRNA and protein expression in PANC-1 cells. These data are similar to results obtained in scleroderma fibroblasts in which CCN2 overexpression was TGFβ/Smad-independent and Sp1/BCE-1-dependent (Holmes et al. 2001, 2003; Chen et al. 2006). These results strongly support the notion that TGFβ is not required under situations when CCN2 is constitutively expressed. Previously, we have shown that the Ras/MEK/ERK pathway regulates CCN2 levels in response to TGF-β and ET-1 (Chen et al. 2002; Leask et al. 2003; Shi-wen et al. 2004). Indeed, it is interesting to note that antagonizing receptors for either TGFβ or ET-1 was not effective at blocking CCN2 expression in PANC-1 cells (data not shown), in spite of the fact that ET-1 induces the CCN2 promoter via BCE-1 and ras/MEK/ERK in lung fibroblasts (Shi-wen et al. 2004). However, this is the first report examining the mechanism underlying MEK/ERK-dependent regulation of CCN2 expression. The identity of the transcription factor(s) binding to BCE-1, an element initially identified by Grotendorst et al. (1996), has not thus far been identified.
The involvement of ras/MEK/ERK in constitutively elevated CCN2 expression in pathological states has not until now been demonstrated. It is interesting to note that ê-Ras, a member of the ras family of GTP-binding proteins, has been found to be mutated in pancreatic cancer, decreasing its GTP hydrolysis, causing it to be constitutively active, constantly activating the MEK/ERK pathway, leading to extensive cell proliferation (Hezel et al. 2006) in a fashion that is likely to be independent of growth factors and cytokines. This observation is consistent with the fact that antagonising TGFβ or ET-1 receptors did not affect the CCN2 promoter activity in PANC-1 cells. Thus, CCN2 is likely to be operating downstream of hyperactive ras in promoting oncogenesis. Previous studies using an anti-CCN2 antibody have suggested that anti-CCN2 strategies may be useful in combating pancreatic cancer (Aikawa et al. 2006; Dornhofer et al. 2006); however, in these studies no mechanistic insight underlying the effect of the antibody was provided. Exogenous CCN2 has been shown to promote angiogenesis, proliferation and migration in a context-specific fashion (Leask and Abraham 2006). However, the effect of endogenous CCN2 in cancer, including that of the pancreas is unclear, and awaits further studies.
References
Abraham DJ, Shiwen X, Black CM, Sa S, Xu Y, Leask A (2000) Tumor necrosis factor alpha suppresses the induction of connective tissue growth factor by transforming growth factor-beta in normal and scleroderma fibroblasts. J Biol Chem 275:15220–15225
Aikawa T, Gunn J, Spong SM, Klaus SJ, Korc M (2006) Connective tissue growth factor-specific antibody attenuates tumor growth,metastasis, and angiogenesis in an orthotopic mouse model of pancreatic cancer. Mol Cancer Ther 5:1108–1116
Babic AM, Chen CC, Lau LF (1999) Fisp12/mouse connective tissue growth factor mediates endothelial cell adhesion and migration through integrin alphavbeta3, promotes endothelial cell survival, and induces angiogenesis in vivo. Mol Cell Biol 19:2958–2966
Blom IE, Goldschmeding R, Leask A (2001) Gene regulation of connective tissue growth factor: new targets for antifibrotic therapy? Matrix Biol 21:473–482
Chen Y, Blom IE, Sa S, Goldschmeding R, Abraham DJ, Leask A (2002) CTGF expression in mesangial cells: involvement of SMADs, MAP kinase, and PKC. Kidney Int 62:149–159
Chen Y, Abraham DJ, Shi-Wen X, Pearson JD, Black CM, Lyons KM, Leask A (2004) CCN2 (connective tissue growth factor) promotes fibroblast adhesion to fibronectin. Mol Biol Cell 15:5635–5646
Chen Y, Shi-wen X, Eastwood M, Black CM, Denton CP, Leask A, Abraham DJ (2006) Contribution of activin receptor-like kinase 5 (transforming growth factor beta receptor type I) signaling to the fibrotic phenotype of scleroderma fibroblasts. Arthritis Rheum 54:1309–1316
Dornhofer N, Spong S, Bennewith K, Salim A, Klaus S, Kambham N, Wong C, Kaper F, Sutphin P, Nacamuli R, Hockel M, Le Q, Longaker M, Yang G, Koong A, Giaccia A (2006) Connective tissue growth factor-specific monoclonal antibody therapy inhibits pancreatic tumor growth and metastasis. Cancer Res 66:5816–5827
Grotendorst GR, Okochi H, Hayashi N (1996) A novel transforming growth factor beta response element controls the expression of the connective tissue growth factor gene. Cell Growth Differ 7:469–480
Hartel M, Di Mola FF, Gardini A, Zimmermann A, Di Sebastiano P, Guweidhi A, Innocenti P, Giese T, Giese N, Buchler MW, Friess H (2004) Desmoplastic reaction influences pancreatic cancer growth behavior. World J Surg 28:818–825
Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA (2006) Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev 20:1218–1249
Holmes A, Abraham DJ, Sa S, Shiwen X, Black CM, Leask A (2001) CTGF and SMADs, maintenance of scleroderma phenotype is independent of SMAD signaling. J Biol Chem 276:10594–10601
Holmes A, Abraham DJ, Chen Y, Denton C, Shi-wen X, Black CM, Leask A (2003) Constitutive connective tissue growth factor expression in scleroderma fibroblasts is dependent on Sp1. J Biol Chem 278:41728–41733
Kennedy L, Liu S, Shi-Wen X, Chen Y, Eastwood M, Carter DE, Lyons KM, Black CM, Abraham DJ, Leask A (2007) CCN2 is necessary for the function of mouse embryonic fibroblasts. Exp Cell Res 313:952–964
Leask A, Abraham DJ (2006) All in the CCN family: essential matricellular signaling modulators emerge from the bunker. J Cell Sci 119:4803–4810
Leask A, Holmes A, Black CM, Abraham DJ (2003) Connective tissue growth factor gene regulation. Requirements for its induction by transforming growth factor-beta 2 in fibroblasts. J Biol Chem 278:13008–13015
Shi-wen X, Howat SL, Renzoni EA, Holmes A, Pearson JD, Dashwood MR, Bou-Gharios G, Denton CP, du Bois RM, Black CM, Leask A, Abraham DJ (2004) Endothelin-1 induces expression of matrix-associated genes in lung fibroblasts through MEK/ERK. J Biol Chem 279:23098–23103
Shi-wen X, Stanton LA, Kennedy L, Pala D, Chen Y, Howat SL, Renzoni EA, Carter DE, Bou-Gharios G, Stratton RJ, Pearson JD, Beier F, Lyons KM, Black CM, Abraham DJ, Leask A (2006) CCN2 is necessary for adhesive responses to transforming growth factor-beta1 in embryonic fibroblasts. J Biol Chem 281:10715–10726
Stratton R, Rajkumar V, Ponticos M, Nichols B, Shiwen X, Black CM, Abraham DJ, Leask A (2002) Prostacyclin derivatives prevent the fibrotic response to TGFbeta by inhibiting the ras/MEK/ERK pathway. FASEB J 16:1949–1951
van Beek JP, Kennedy L, Rockel JS, Bernier SM, Leask A (2006) The induction of CCN2 by TGFbeta1 involves Ets-1. Arthritis Res Ther 8:R36
Wenger C, Ellenrieder V, Alber B, Lacher U, Menke A, Hameister H, Wilda M, Iwamura T, Beger HG, Adler G, Gress TM (1999) Expression and differential regulation of connective tissue growth factor in pancreatic cancer cells. Oncogene 18:1073–1080
Yamamoto T, Taya S, Kaibuchi K (1999) Ras-induced transformation and signaling pathway. J Biochem (Tokyo) 126:799–803
Acknowledgements
Our work is supported by grants from the Canadian Institutes of Health Research and the Canadian Foundation for Innovation. A.L. is a New Investigator of the Arthritis Society (Scleroderma Society of Ontario) and a recipient of an Early Researcher Award. M.P. performed this work as partial fulfillment of an Honors Bachelor of Science degree from the University of Western Ontario.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Pickles, M., Leask, A. Analysis of CCN2 promoter activity in PANC-1 cells: regulation by ras/MEK/ERK. J. Cell Commun. Signal. 1, 85–90 (2007). https://doi.org/10.1007/s12079-007-0008-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12079-007-0008-9