Abstract
The basic helix-loop-helix (bHLH) transcription factor family, which is widely prevalent in higher plants, plays an important role in regulating plant growth and development under various environmental factors. However, there is still no genome-wide analysis of the bHLH family conducted in mango (Mangifera indica L.). In this study, 212 MibHLH genes were identified in the mango genome by genome wide-analysis and they are unequally distributed across all the mango chromosomes. Based on the phylogenetic tree analysis, MibHLH proteins were divided into 26 subgroups. GO and protein-protein interaction network analyses showed bHLH protein family has multiple biological functions in mango. Several cis-elements related to hormone responses such as CGTCA-motif and TGACG-motif, ABRE, GARE-motif, TCA-element and TGA-element were identified in the upstream regions of MibHLH genes. The results provide some valuable information for exploring the functional roles of the MibHLH genes in mango.





Similar content being viewed by others
Data Availability
All related data are available within the manuscript and its additional files.
References
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME suite: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208. https://doi.org/10.1093/nar/gkp335
Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39:366–380. https://doi.org/10.1111/j.1365-313X.2004.02138.x
Ben-Simhon Z, Judeinstein S, Nadler-Hassar T et al (2011) A pomegranate (Punica granatum L.) WD40-repeat gene is a functional homologue of Arabidopsis TTG1 and is involved in the regulation of anthocyanin biosynthesis during pomegranate fruit development. Planta. https://doi.org/10.1007/s00425-011-1438-4
Breuninger H, Thamm A, Streubel S, Sakayama H, Nishiyama T, Dolan L (2016) Diversification of a transcription factor family led to the evolution of antagonistically acting genetic regulators of root hair growth. Curr Biol 26:1622–1628. https://doi.org/10.1016/j.cub.2016.04.060
Buck MJ, Atchley WR (2003) Phylogenetic analysis of plant basic helix-loop-helix proteins. J Mol Evol 56:742–750. https://doi.org/10.1007/s00239-002-2449-3
Dai Z, An K, Edward GE, An G (1999) Functional role of CAAT box element of the nopaline synthase (nos) promoter. J Plant Biol 42:181–185. https://doi.org/10.1007/BF03031028
Deng C, Ye H, Fan M, Pu T, Yan J (2017) The rice transcription factors OsICE confer enhanced cold tolerance in transgenic Arabidopsis. Plant Signal Behav 12:e1316442. https://doi.org/10.1080/15592324.2017.1316442
Dubos C, Le Gourrierec J, Baudry A et al (2008) MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J 55:940–953. https://doi.org/10.1111/j.1365-313X.2008.03564.x
Feller A, MacHemer K, Braun EL, Grotewold E (2011) Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J 66:94–116. https://doi.org/10.1111/j.1365-313X.2010.04459.x
Feng Y, Xu P, Li B, Li P, Wen X, An F, Gong Y, Xin Y, Zhu Z, Wang Y, Guo H (2017) Ethylene promotes root hair growth through coordinated EIN3/EIL1 and RHD6/RSL1 activity in Arabidopsis. Proc Natl Acad Sci U S A 114:13834–13839. https://doi.org/10.1073/pnas.1711723115
Ferguson AC, Pearce S, Band LR, Yang C, Ferjentsikova I, King J, Yuan Z, Zhang D, Wilson ZA (2017) Biphasic regulation of the transcription factor ABORTED MICROSPORES (AMS) is essential for tapetum and pollen development in Arabidopsis. New Phytol 213:778–790. https://doi.org/10.1111/nph.14200
Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, Pauwels L, Witters E, Puga MI, Paz-Ares J, Goossens A, Reymond P, de Jaeger G, Solano R (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23:701–715. https://doi.org/10.1105/tpc.110.080788
Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K (2011) ABA-mediated transcriptional regulation in response to osmotic stress in plants. J Plant Res 124:509–525
Gao F, Robe K, Gaymard F, et al (2019) The transcriptional control of iron homeostasis in plants: a tale of bHLH transcription factors? Front. Plant Sci, p 18. https://doi.org/10.3389/fpls.2019.00006
Geng Y, Wu R, Wee CW, Xie F, Wei X, Chan PMY, Tham C, Duan L, Dinneny JR (2013) A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell 25:2132–2154. https://doi.org/10.1105/tpc.113.112896
Girin T, Paicu T, Stephenson P, Fuentes S, Körner E, O’Brien M, Sorefan K, Wood TA, Balanzá V, Ferrándiz C, Smyth DR, Østergaard L (2011) INDEHISCENT and SPATULA interact to specify carpel and valve margin tissue and thus promote seed dispersal in Arabidopsis. Plant Cell 23:3641–3653. https://doi.org/10.1105/tpc.111.090944
Guo XJ, Wang JR (2017) Global identification, structural analysis and expression characterization of bHLH transcription factors in wheat. BMC Plant Biol 17:90. https://doi.org/10.1186/s12870-017-1038-y
Hichri I, Heppel SC, Pillet J, Léon C, Czemmel S, Delrot S, Lauvergeat V, Bogs J (2010) The basic helix-loop-helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine. Mol Plant 3:509–523. https://doi.org/10.1093/mp/ssp118
Ito S, Song YH, Josephson-Day AR, Miller RJ, Breton G, Olmstead RG, Imaizumi T (2012) FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc Natl Acad Sci U S A 109:3582–3587. https://doi.org/10.1073/pnas.1118876109
Jahurul MHA, Zaidul ISM, Ghafoor K, et al (2015) Mango (Mangifera indica L.) by-products and their valuable components: a review. Food Chem, pp 173–180.https://doi.org/10.1016/j.foodchem.2015.03.046
Jiang Y, Yang B, Deyholos MK (2009) Functional characterization of the Arabidopsis bHLH92 transcription factor in abiotic stress. Mol Gen Genomics 282:503–516. https://doi.org/10.1007/s00438-009-0481-3
Jones S (2004) An overview of the basic helix-loop-helix proteins. Genome Biol 5:226
Karas B, Amyot L, Johansen C, Sato S, Tabata S, Kawaguchi M, Szczyglowski K (2009) Conservation of Lotus and Arabidopsis basic helix-loop-helix proteins reveals new players in root hair development. Plant Physiol 151:1175–1185. https://doi.org/10.1104/pp.109.143867
Kavas M, Baloğlu MC, Atabay ES, Ziplar UT, Daşgan HY, Ünver T (2015) Genome-wide characterization and expression analysis of common bean bHLH transcription factors in response to excess salt concentration. Mol Gen Genomics 291:129–143. https://doi.org/10.1007/s00438-015-1095-6
Khanna R, Shen Y, Marion CM, Tsuchisaka A, Theologis A, Schäfer E, Quail PH (2007) The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell 19:3915–3929. https://doi.org/10.1105/tpc.107.051508
Kim JA, Yun J, Lee M, et al (2005) A basic helix-loop-helix transcription factor regulates cell elongation and seed germination. Mol Cells 19(3):334–41. http://europepmc.org/abstract/MED/15995349
Le Hir R, Castelain M, Chakraborti D et al (2017) AtbHLH68 transcription factor contributes to the regulation of ABA homeostasis and drought stress tolerance in Arabidopsis thaliana. Physiol Plant 160:312–327. https://doi.org/10.1111/ppl.12549
Ledent V, Paquet O, Vervoort M (2002) Phylogenetic analysis of the human basic helix-loop-helix proteins. Genome Biol 3:research0030.1. https://doi.org/10.1186/gb-2002-3-6-research0030
Ledent V, Vervoort M (2001) The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Res 11:754–770
Li C, Qiu J, Huang S, et al (2019) AaMYB3 interacts with AabHLH1 to regulate proanthocyanidin accumulation in Anthurium andraeanum (Hort.)—another strategy to modulate pigmentation. Hortic Res 1:6–14. https://doi.org/10.1038/s41438-018-0102-6
Li X, Duan X, Jiang H, Sun Y, Tang Y, Yuan Z, Guo J, Liang W, Chen L, Yin J, Ma H, Wang J, Zhang D (2006) Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol 141:1167–1184. https://doi.org/10.1104/pp.106.080580
Liu X, Cao X, Shi S, Zhao N, Li D, Fang P, Chen X, Qi W, Zhang Z (2018) Comparative RNA-Seq analysis reveals a critical role for brassinosteroids in rose (Rosa hybrida) petal defense against Botrytis cinerea infection. BMC Genet 19:62. https://doi.org/10.1186/s12863-018-0668-x
Meier-Andrejszki L, Bjelić S, Naud JF, Lavigne P, Jelesarov I (2007) Thermodynamics of b-HLH-LZ protein binding to DNA: the energetic importance of protein-DNA contacts in site-specific E-box recognition by the complete gene product of the max p21 transcription factor. Biochemistry. 46:12427–12440. https://doi.org/10.1021/bi701081q
Moon J, Zhu L, Shen H, Huq E (2008) PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc Natl Acad Sci U S A 105:9433–9438. https://doi.org/10.1073/pnas.0803611105
Naval MM, Gil-Munoz F, Lloret A, et al (2017) Molecular characterization of a TTG1-like gene expressed in persimmon fruit. In: Acta Horticulturae 1172. https://doi.org/10.17660/ActaHortic.2017.1172.67
Oh E, Kang H, Yamaguchi S, Park J, Lee D, Kamiya Y, Choi G (2009) Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in arabidopsis. Plant Cell 21:403–419. https://doi.org/10.1105/tpc.108.064691
Oh E, Kim J, Park E, Kim JI, Kang C, Choi G (2004) PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16:3045–3058. https://doi.org/10.1105/tpc.104.025163
Oh E, Yamaguchi S, Hu J, Yusuke J, Jung B, Paik I, Lee HS, Sun TP, Kamiya Y, Choi G (2007) PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19:1192–1208. https://doi.org/10.1105/tpc.107.050153
Ohashi-Ito K, Matsukawa M, Fukuda H (2013) An atypical bHLH transcription factor regulates early xylem development downstream of auxin. Plant Cell Physiol 54:398–405. https://doi.org/10.1093/pcp/pct013
Pires N, Dolan L (2010) Origin and diversification of basic-helix-loop-helix proteins in plants. Mol Biol Evol 27:862–874. https://doi.org/10.1093/molbev/msp288
Ptashne M (1988) How eukaryotic transcriptional activators work. Nature 335:683–689
Qi X, Torii KU (2018) Hormonal and environmental signals guiding stomatal development. BMC Biol 16
Ramsay NA, Glover BJ (2005) MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci 10:63–70
Ribeiro SMR, Schieber A (2010) Bioactive compounds in mango (Mangifera indica L.). Bioactive foods in promoting health, pp 507–523. https://doi.org/10.1016/B978-0-12-374628-3.00034-7
Salih H, He S, Li H, Peng Z, du X (2020) Investigation of the EIL/EIN3 transcription factor gene family members and their expression levels in the early stage of cotton Fiber development. Plants 9:128. https://doi.org/10.3390/plants9010128
Salih H, Odongo MR, Gong W, He S, du X (2019) Genome-wide analysis of cotton C2H2-zinc finger transcription factor family and their expression analysis during fiber development. BMC Plant Biol 19:400. https://doi.org/10.1186/s12870-019-2003-8
Schuster C, Gaillochet C, Lohmann JU (2015) Arabidopsis HECATE genes function in phytohormone control during gynoecium development. Dev. 142:3343–3350. https://doi.org/10.1242/dev.120444
Shan X, Zhang W, Yu F et al (2019) Genome-wide analysis of basic helix–loop–helix superfamily members reveals organization and chilling-responsive patterns in cabbage (Brassica oleracea var. capitata l.). Genes (Basel). https://doi.org/10.3390/genes10110914
Shor E, Paik I, Kangisser S, Green R, Huq E (2017) PHYTOCHROME INTERACTING FACTORS mediate metabolic control of the circadian system in Arabidopsis. New Phytol 215:217–228. https://doi.org/10.1111/nph.14579
Sivakumar D, Jiang Y, Yahia EM (2011) Maintaining mango (Mangifera indica L.) fruit quality during the export chain. Food Res Int. https://doi.org/10.1016/j.foodres.2010.11.022
Sorensen AM, Kröber S, Unte US, Huijser P, Dekker K, Saedler H (2003) The Arabidopsis aborted microspores (ams) gene encodes a MYC class transcription factor. Plant J 33:413–423. https://doi.org/10.1046/j.1365-313X.2003.01644.x
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447–D452. https://doi.org/10.1093/nar/gku1003
Takatsuji H (1999) Zinc-finger proteins: the classical zinc finger emerges in contemporary plant science. Plant Mol Biol 39:1073–1078
Toledo-Ortiz G, Huq E, Quail PH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15:1749–1770. https://doi.org/10.1105/tpc.013839
Verma D, Jalmi SK, Bhagat PK, Verma N, Sinha AK (2019) A bHLH transcription factor, MYC2, imparts salt intolerance by regulating proline biosynthesis in Arabidopsis. FEBS J 287:2560–2576. https://doi.org/10.1111/febs.15157
Vervoort M (2001) The basic Helix-loop-Helix protein family : comparative genomics and phylogenetic analysis derivation of comprehensive sets. Genome Biol. https://doi.org/10.1101/gr.177001.1
Wang C, Yao X, Yu D, Liang G (2017) Fe-deficiency-induced expression of bHLH104 enhances Fe-deficiency tolerance of Arabidopsis thaliana. Planta 246:421–431. https://doi.org/10.1007/s00425-017-2703-y
Wang GL, Jiang BH, Rue EA, Semenza GL (1995) Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A 92:5510–5514. https://doi.org/10.1073/pnas.92.12.5510
Wang P et al (2020) The genome evolution and domestication of tropical fruit mango. Genome Biol 21:60. https://doi.org/10.1186/s13059-020-01959-8
Wang Q, Guan Y, Wu Y, Chen H, Chen F, Chu C (2008) Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Mol Biol 67:589–602. https://doi.org/10.1007/s11103-008-9340-6
Wang R, Zhao P, Kong N, Lu R, Pei Y, Huang C, Ma H, Chen Q (2018) Genome-wide identification and characterization of the potato bHLH transcription factor family. Genes (Basel) 9:54. https://doi.org/10.3390/genes9010054
Waseem M, Rong X, Li Z (2019) Dissecting the role of a basic helix-loop-helix transcription factor, SlBHLH22, under salt and drought stresses in transgenic Solanum lycopersicum L. Front Plant Sci 10:734. https://doi.org/10.3389/fpls.2019.00734
Yahia EM (2011) Mango (Mangifera indica L.). in: postharvest biology and Technology of Tropical and Subtropical Fruits: Cocona to mango. Food Science, Technology and Nutrition 492-565:566e–567e. https://doi.org/10.1533/9780857092885.492
Yan P, Zeng Y, Shen W et al (2020) Nimble Cloning: A Simple, Versatile, and Efficient System for Standardized Molecular Cloning. Front Bioeng Biotechnol. https://doi.org/10.3389/fbioe.2019.00460
Yu MH, Zhao ZZ, He JX (2018) Brassinosteroid signaling in plant–microbe interactions. Int J Mol Sci 19:4091. https://doi.org/10.3390/ijms19124091
Zarka DG, Vogel JT, Cook D, Thomashow MF (2003) Cold induction of Arabidopsis CBF genes involves multiple ICE (inducer of CBF expression) promoter elements and a cold-regulatory circuit that is desensitized by low temperature. Plant Physiol 133:910–918. https://doi.org/10.1104/pp.103.027169
Zhang T, Mo J, Zhou K, Chang Y, Liu Z (2018) Overexpression of Brassica campestris BcICE1 gene increases abiotic stress tolerance in tobacco. Plant Physiol Biochem 132:515–523. https://doi.org/10.1016/j.plaphy.2018.09.039
Zhao K, Li S, Yao W, Zhou B, Li R, Jiang T (2018) Characterization of the basic helix-loop- helix gene family and its tissue-differential expression in response to salt stress in poplar. PeerJ. 6:e4502. https://doi.org/10.7717/peerj.4502
Zhao M, Morohashi K, Hatlestad G, Grotewold E, Lloyd A (2008) The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 135:1991–1999. https://doi.org/10.1242/dev.016873
Zhu E, You C, Wang S, Cui J, Niu B, Wang Y, Qi J, Ma H, Chang F (2015) The DYT1-interacting proteins bHLH010, bHLH089 and bHLH091 are redundantly required for Arabidopsis anther development and transcriptome. Plant J 83:976–990. https://doi.org/10.1111/tpj.12942
Zhu J, Chen H, Li H, Gao JF, Jiang H, Wang C, Guan YF, Yang ZN (2008) Defective in Tapetal development and function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J 55:266–277. https://doi.org/10.1111/j.1365-313X.2008.03500.x
Funding
The authors appreciate the support of the Central Public-interest Scientific Institution BasalResearch Fund for Chinese Academy of Tropical Agricultural Sciences, grant number (No1630092020012)and the earmarked fund for the Belt and Road Tropical Project(BARTP-07).
Author information
Authors and Affiliations
Contributions
HS and NNWH designed the experiment. HS analyzed the results and prepared the manuscript. LT and RZ and revised the manuscript. All authorsreviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Communicated by: Yuan Qin
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Figure S1
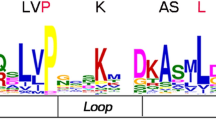
Phylogenetic tree analysis, conserved motifs MibHLH protein family (A) the phylogenetic tree of bHLH proteins in mango (B) Ten motifs MibHLH proteins were identified by MEME are represented by colored boxes and consensus sequences. (PDF 279 kb)
Table S1
Primer sequence lists and nimble cloning method were used in this study. (DOCX 18 kb)
Table S2
MibHLH members, amino acids length, sub-molecular localization, isoelectric point (pI) and molecular weight. (XLSX 18 kb)
Table S3
Chromosomal position, gene annotation gene length, Exon/Intron of bHLH genes in mango genome. The positive (+) and negative (−) symbols following each gene represent forward and reverse orientations, respectively, on the chromosome. (XLSX 24 kb)
Table S4
Most abundant cis-regulatory elements were detected in promoter sites of MibHLH genes. (XLSX 246 kb)
Table S5
Predicted functions of the MibHLHs with the function of their homologs verified in Arabidopsis by phylogenetic analysis. (XLSX 18 kb)
Table S6
Summary information Of 212 MibHLH proteins was found in STRING database based on their homologs from Arabidopsis. (XLSX 27 kb)
Rights and permissions
About this article
Cite this article
Salih, H., Tan, L. & Htet, N.N.W. Genome-Wide Identification, Characterization of bHLH Transcription Factors in Mango. Tropical Plant Biol. 14, 72–81 (2021). https://doi.org/10.1007/s12042-020-09277-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12042-020-09277-w