Abstract
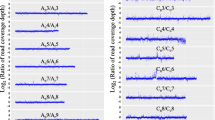
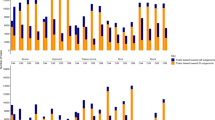
The aneuploidy chromosome addition lines with individual chromosomes of one species into the whole genome of another species developed from interspecific or intergeneric crosses are ideal for studying the gene expression regulation under asymmetrical genome interactions. Here, we chose two rapeseed-radish addition lines (2n=40, AACC+2R) with all chromosomes of rapeseed and one pair of radish chromosomes carrying 25S rDNA loci and both 5S and 25S rDNA loci, respectively. Transcriptome sequencing of these two rapeseed-radish addition lines, together with two parents (Brassica napus, 2n=38, AACC; Raphanus sativus, 2n=18, RR), was performed to assess gene expression changes due to the aneuploidy effect. Our results showed that the global gene expression perturbations in two addition lines showed asymmetric distributions between A and C subgenomes, for more downregulated genes located in the C subgenome. Moreover, several dysregulated domains occurred in the addition lines and majority of them were clustered in C subgenome, further revealing that C subgenome was more inclined to be repressed by the aneuploidy. Homoeolog expression was slightly biased toward C subgenome in the parent B. napus, but had no preference to either of A or C subgenome in the addition lines. The total number of biased genes increased sharply and most of them were shared only by two addition lines, indicating that aneuploidy could change the extent and direction of homoeolog expression. The triplicated subgenomes which were orthologs to the chromosomes of Arabidopsis thaliana also exhibited dramatic alternations in two additions. Together, our results revealed that the addition of individual radish chromosomes could induce dramatically transcriptomic disturbances and those expression changes gave asymmetric distributions between two subgenomes in B. napus. The possible mechanisms including the uniparental expression of rRNA genes in the allopolyploids and additions for the findings are discussed.






Similar content being viewed by others
References
Budahn H., Schrader O. and Peterka H. 2007 Development of a complete set of disomic rape-radish chromosome addition lines. Euphytica 162, 117–128.
Chalhoub B., Denoeud F., Liu S., Parkin I. A., Tang H., Wang X. et al. 2014 Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953.
Chang S. B. and de Jong H. 2005 Production of alien chromosome additions and their utility in plant genetics. Cytogenet. Genome Res. 109, 335–343.
Chen Z. J. 2007 Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Ann. Rev. Plant Biol. 58, 377–406.
Cheng F., Sun R., Hou X., Zheng H., Zhang F., Zhang Y. et al. 2016 Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nat. Genet. 48, 1218–1224.
Cho S., Garvin D. F. and Muehlbauer G. J. 2006 Transcriptome analysis and physical mapping of barley genes in wheat-barley chromosome addition lines. Genetics 172, 1277–1285.
Ding L., Zhao Z. G., Ge X. H. and Li Z. Y. 2013 Intergeneric addition and substitution of Brassica napus with different chromosomes from Orychophragmus violaceus: Phenotype and cytology. Sci. Horticult. 164, 303–309.
Dong Z. B., Yu J., Li H., Huang W., Xu L., Zhao Y. et al. 2018 Transcriptional and epigenetic adaptation of maize chromosomes in oat-maize addition lines. Nucleic Acids Res. 46, 5012–5028.
Du X. Z., Ge X. H., Yao X. C., Zhao Z. G. and Li Z. Y. 2009 Production and cytogenetic characterization of intertribal somatic hybrids between Brassica napus and Isatis indigotica and backcross progenies. Plant Cell Rep. 28, 1105–1113.
Fu W. Q., Zhao Z. G., Ge X. H., Ding L. and Li Z. Y. 2014 Anatomy and transcript profiling of gynoecium development in female sterile Brassica napus mediated by one alien chromosome from Orychophragmus violaceus. BMC Genomics 15, 61.
Gao L., Diarso M., Zhang A., Zhang H., Dong Y., Liu L. et al. 2016 Heritable alteration of DNA methylation induced by whole-chromosome aneuploidy in wheat. New Phytol. 209, 364–375.
Gautam M., Dang Y., Ge X., Shao Y. and Li Z. 2016 Genetic and epigenetic changes in oilseed rape (Brassica napus L.) extracted from intergeneric allopolyploid and additions with Orychophragmus. Front. Plant Sci. 7, 438.
Ge X. H., Ding L. and Li Z. Y. 2013 Nucleolar dominance and different genome behaviors in hybrids and allopolyploids. Plant Cell Rep. 32, 1661–1673.
Ge X. H., Wang J. and Li Z. Y. 2009 Different genome-specific chromosome stabilities in synthetic Brassica allohexaploids revealed by wide crosses with Orychophragmus. Ann. Bot. 104, 19–31.
Gu X. and Lu T. 2016 Commentary: Comparative transcriptome analysis of Raphanus sativus tissues. Front. Plant Sci. 6, 1191.
Huettel B., Kreil D. P., Matzke M. and Matzke A. J. 2008 Effects of aneuploidy on genome structure, expression, and interphase organization in Arabidopsis thaliana. PLoS Genet. 4, e1000226.
Inaba R. and Nishio T. 2002 Phylogenetic analysis of Brassiceae based on the nucleotide sequences of the S-locus related gene, SLR1. Theor. Appl. Genet. 105, 1159–1165.
Jeong Y. M., Kim N., Ahn B. O., Oh M., Chung W. H., Chung H. et al. 2016 Elucidating the triplicated ancestral genome structure of radish based on chromosome-level comparison with the Brassica genomes. Theor. Appl. Genet. 129, 1357–1372.
Kang L., Du X., Zhou Y., Zhu B., Ge X. and Li Z. 2014 Development of a complete set of monosomic alien addition lines between Brassica napus and Isatis indigotica (Chinese woad). Plant Cell Rep. 33, 1355–1364.
Letourneau A., Santoni F. A., Bonilla X., Sailani M. R., Gonzalez D., Kind J. et al. 2014 Domains of genome-wide gene expression dysregulation in Down’s syndrome. Nature 508, 345–350.
Li F., Hasegawa Y., Saito M., Shirasawa S., Fukushima A., Ito T. et al. 2011 Extensive chromosome homoeology among Brassiceae species were revealed by comparative genetic mapping with high-density EST-based SNP markers in radish (Raphanus sativus L.). DNA Res. 18, 401–411.
Li Z., Liu H. L. and Luo P. 1995 Production and cytogenetics of intergeneric hybrids between Brassica napus and Orychophragmus violaceus. Theor. Appl. Genet. 91, 131–136.
Liu S., Liu Y., Yang X., Tong C., Edwards D., Parkin I. A. et al. 2014 The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 5, 3930.
Mitsui Y., Shimomura M., Komatsu K., Namiki N., Shibata-Hatta M., Imai M. et al. 2015 The radish genome and comprehensive gene expression profile of tuberous root formation and development. Sci. Rep. 5, 10835.
Moghe G. D., Hufnagel D. E., Tang H., Xiao Y., Dworkin I., Town C. D. et al. 2014 Consequences of whole-genome triplication as revealed by comparative genomic analyses of the wild radish Raphanus raphanistrum and three other Brassicaceae species. Plant Cell 26, 1925–1937.
Mun J. H., Chung H., Chung W. H., Oh M., Jeong Y. M., Kim N. et al. 2015 Construction of a reference genetic map of Raphanus sativus based on genotyping by whole-genome resequencing. Theor. Appl. Genet. 128, 259–272.
Pikaard C. S. 2000 Nucleolar dominance: uniparental gene silencing on a multi-megabase scale in genetic hybrids. Plant Mol. Biol. 43, 163.
Prakash S., Bhat S. R., Quiros C. F., Kirti P. B. and Chopra V. L. 2009 Brassica and its close allies: cytogenetics and evolution. Plant Breed. Rev. 31, 21–187.
Rey E., Abrouk M., Keeble-Gagnère G., Karafiátová M., Vrána J., Balzergue S. et al. 2018 Transcriptome reprogramming due to the introduction of a barley telosome into bread wheat affects more barley genes than wheat. Plant Biotechnol. J. 16, 1767–1777.
Sears E. R. 1954 The aneuploids of common wheat. Agric. Exp. State Res. Bull 572, 1–58.
Siegel J. J. and Amon A. 2012 New insights into the troubles of aneuploidy. Annu. Rev. Cell Dev. Biol. 28, 189–214.
Soltis P. S., Soltis D. E. 2012 Polyploidy and genome evolution, pp. 1–416. Springer-Verlag, New York,.
Wang X., Wang H., Wang J., Sun R., Wu J., Liu S. et al. 2011 The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1039.
Wang L., Wei L., Wang L. and Xu C. 2002 Effects of peroxidase on hyperlipidemia in mice. J. Agri. Food Chem. 50, 868.
Wang Y., Xu L., Chen Y. L., Shen H., Gong Y., Limera C. et al. 2013 Transcriptome profiling of radish (Raphanus sativus L.) root and identification of genes involved in response to lead (Pb) stress with next generation sequencing. PLoS One 8, e66539.
Warwick S. I. and Black L. D. 1991 Molecular systematics of Brassica and allied genera (Subtribe Brassicinae, Brassiceae) —chloroplast genome and cytodeme congruence. Theor. Appl. Genet. 82, 81–92.
Wu G., Zhang L., Yin Y., Wu J., Yu L., Zhou Y. et al. 2015 Sequencing, de novo assembly and comparative analysis of Raphanus sativus transcriptome. Front. Plant Sci. 6, 198.
Yamasaki M., Omi Y., Fujii N., Ozaki A., Nakama A., Sakakibara Y. et al. 2009 Mustard oil in “Shibori Daikon” a variety of Japanese radish, selectively inhibits the proliferation of H-ras-transformed 3Y1 cells. Biosci. Biotechnol. Biochem. 73, 2217–2221.
Yang Y. W., Tai P. Y., Chen Y. and Li W. H. 2002 A study of the phylogeny of Brassica rapa, B. nigra, Raphanus sativus, and their related genera using noncoding regions of chloroplast DNA. Mol. Phylogenet. Evol. 23, 268–275.
Yoo M. J., Szadkowski E. and Wendel J. F. 2013 Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity 110, 171–180.
Zhang A., Li N., Gong L., Gou X., Wang B., Deng X. et al. 2017 Global analysis of gene expression in response to whole-chromosome aneuploidy in hexaploid wheat. Plant Physiol. 175, 828–847.
Zhang D., Pan Q., Cui C., Zhu B., Ge X., Shao Y. et al. 2016 Genome-wide gene expressions respond differently to A-subgenome origins in Brassica napus synthetic hybrids and natural allotetraploid. Front. Plant Sci. 7, 1508.
Zhang D., Pan Q., Tan C., Liu L., Ge X., Li Z. et al. 2018 Homoeolog expression is modulated differently by different subgenomes in Brassica napus hybrids and allotetraploids. Plant Mol. Biol. Rep. 36, 387–398.
Zhang L. B., Jia H. B., Ying Y. T., Wu G., Xia H., Wang X. et al. 2013 Transcriptome analysis of leaf tissue of Raphanus sativus by RNA sequencing. PLoS One 8, e80350.
Zhang L. B., Ma C., Chao H. B., Long Y., Wu J., Li Z. et al. 2019 Integration of metabolome and transcriptome reveals flavonoid accumulation in the intergeneric hybrid between Brassica rapa and Raphanus sativus. Sci. Rep. 9, 18368.
Zhang Y., Malone J. H., Powell S. K., Periwal V., Spana E., Macalpine D. M. et al. 2010 Expression in aneuploidy Drosophila S2 cells. PLoS Biol. 8, 403.
Zhu B., Shao Y., Pan Q., Ge X. and Li Z. 2015 Genome-wide gene expression perturbation induced by loss of C2 chromosome in allotetraploid Brassica napus L. Front. Plant Sci. 6, 763.
Zhu B., Tu Y. Q., Zeng P., Ge X. and Li Z. 2016 Extraction of the constituent subgenomes of the natural allopolyploid rapeseed (Brassica napus L.). Genetics 204, 1015–1027.
Zhu B., Xiang Y., Zeng P., Cai B., Huang X., Ge X. et al. 2018 Genome-wide gene expression disturbance by single A1/C1 chromosome substitution in Brassica rapa restituted from natural B. napus. Front. Plant Sci. 9, 377.
Zhu B., Pan Q., Huo D. G., Cai B., Ge X. and Li Z. 2019 Transcriptional aneuploidy responses of Brassica rapa-oleracea monosomic alien addition lines (MAALs) derived from natural allopolyploid B. napus. Front. Genet. 10, 67.
Acknowledgements
This work was supported by the grants from Hubei Provincial (grant no. 2019CFC907), National Natural Science Foundation of China (grant no. 31701462 and U19A2029) and Open Project of Key Laboratory of Biology and Genetic Improvement of Oil Crops, Ministry of Agriculture and Rural Affairs, China (KF2020011).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Corresponding editor: Manoj Prasad
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shao, Y., Pan, Q., Zhang, D. et al. Global gene expression perturbations in rapeseed due to the introduction of alien radish chromosomes. J Genet 100, 25 (2021). https://doi.org/10.1007/s12041-021-01276-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12041-021-01276-4