Abstract
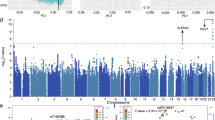
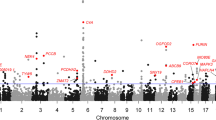
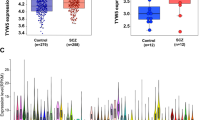
Multiple integrative studies have been performed to identify the potential target genes of the non-coding schizophrenia (SCZ) risk variants. However, all the integrative studies used expression quantitative trait loci (eQTL) data from bulk tissues. Considering the cell type–specific regulatory effect of many genetic variants, it is important to conduct integrative studies using cell type–specific eQTL data. Here, we conduct a Mendelian randomization (MR) study by integrating genome-wide associations of SCZ (74,776 cases and 101,023 controls) and eQTL data (N = 215) from dopaminergic neurons, which were differentiated from human-induced pluripotent stem cell (iPSC) lines. For eQTL from young post-mitotic dopaminergic neurons (differentiation of iPSC for 30 days, D30), we identified 34 genes whose genetically regulated expression in dopaminergic neurons may have a causal role in SCZ. Among which, ARL3 showed the most significant associations with SCZ. For eQTL from more mature dopaminergic neurons (D52), we identified 37 potential SCZ causal genes, and ARL3 and GNL3 showed the most significant associations. Only 12 genes showed significant associations with SCZ in both D30 and D52 eQTL datasets, indicating the time point–specific genetic regulatory effects in young post-mitotic dopaminergic neurons and more mature dopaminergic neurons. Comparing the results from dopaminergic neurons with bulk brain tissues prioritized 2 high-confidence risk genes, including DDHD2 and GALNT10. Our study identifies multiple risk genes whose genetically regulated expression in dopaminergic neurons may have a causal role in SCZ. Further mechanistic investigation will provide pivotal insights into SCZ pathophysiology.




Similar content being viewed by others
Data Availability
All data generated or analyzed during this study are included in this published article (and its supplementary information files).
References
Schizophrenia Working Group of the Psychiatric Genomics Consortium (2014) Biological insights from 108 schizophrenia-associated genetic loci. Nat 511(7510):421–427
Li Z, Chen J, Yu H, He L, Xu Y, Zhang D et al (2017) Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat Genet 49(11):1576–1583
Pardinas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N et al (2018) Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet 50(3):381–389
Lam M, Chen CY, Li Z, Martin AR, Bryois J, Ma X et al (2019) Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat Genet 51(12):1670–1678
Trubetskoy V, Pardiñas AF, Qi T, Panagiotaropoulou G, Awasthi S, Bigdeli TB et al (2022) Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nat 604(7906):502–508
Luo XJ, Mattheisen M, Li M, Huang L, Rietschel M, Borglum AD et al (2015) Systematic integration of brain eQTL and GWAS identifies ZNF323 as a novel schizophrenia risk gene and suggests recent positive selection based on compensatory advantage on pulmonary function. Schizophr Bull 41(6):1294–1308
Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BW et al (2016) Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet 48(3):245–252
Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE et al (2016) Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet 48(5):481–487
Mancuso N, Shi H, Goddard P, Kichaev G, Gusev A, Pasaniuc B (2017) Integrating gene expression with summary association statistics to identify genes associated with 30 complex traits. Am J Hum Genet 100(3):473–487
Gusev A, Mancuso N, Won H, Kousi M, Finucane HK, Reshef Y et al (2018) Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat Genet 50(4):538–548
Yang CP, Li X, Wu Y, Shen Q, Zeng Y, Xiong Q et al (2018) Comprehensive integrative analyses identify GLT8D1 and CSNK2B as schizophrenia risk genes. Nat Commun 9(1):838
Wainberg M, Sinnott-Armstrong N, Mancuso N, Barbeira AN, Knowles DA, Golan D et al (2019) Opportunities and challenges for transcriptome-wide association studies. Nat Genet 51(4):592–599
Hall LS, Medway CW, Pain O, Pardinas AF, Rees EG, Escott-Price V et al (2020) A transcriptome-wide association study implicates specific pre- and post-synaptic abnormalities in schizophrenia. Hum Mol Genet 29(1):159–167
Hall LS, Pain O, O’Brien HE, Anney R, Walters JTR, Owen MJ et al (2020) Cis-effects on gene expression in the human prenatal brain associated with genetic risk for neuropsychiatric disorders. Mol Psychiatry 26(6):2082–2088
Wang JY, Li XY, Li HJ, Liu JW, Yao YG, Li M et al (2021) Integrative analyses followed by functional characterization reveal TMEM180 as a schizophrenia risk gene. Schizophr Bull. https://doi.org/10.1093/schbul/sbab032
He X, Fuller CK, Song Y, Meng Q, Zhang B, Yang X et al (2013) Sherlock: detecting gene-disease associations by matching patterns of expression QTL and GWAS. Am J Hum Genet 92(5):667–680
Ma C, Gu C, Huo Y, Li X, Luo XJ (2018) The integrated landscape of causal genes and pathways in schizophrenia. Transl Psychiatry 8(1):67
Huckins LM, Dobbyn A, Ruderfer DM, Hoffman G, Wang W, Pardinas AF et al (2019) Gene expression imputation across multiple brain regions provides insights into schizophrenia risk. Nat Genet 51(4):659–674
Nott A, Holtman IR, Coufal NG, Schlachetzki JCM, Yu M, Hu R et al (2019) Brain cell type-specific enhancer-promoter interactome maps and disease-risk association. Sci 366(6469):1134–1139
Song M, Pebworth MP, Yang X, Abnousi A, Fan C, Wen J et al (2020) Cell-type-specific 3D epigenomes in the developing human cortex. Nat 587(7835):644–649
Aygun N, Elwell AL, Liang D, Lafferty MJ, Cheek KE, Courtney KP et al (2021) Brain-trait-associated variants impact cell-type-specific gene regulation during neurogenesis. Am J Hum Genet 108(9):1647–1668
Davey Smith G, Hemani G (2014) Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet 23(R1):R89-98
Ference BA, Robinson JG, Brook RD, Catapano AL, Chapman MJ, Neff DR et al (2016) Variation in PCSK9 and HMGCR and risk of cardiovascular disease and diabetes. N Engl J Med 375(22):2144–2153
Davies NM, Holmes MV, Davey SG (2018) Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ 12(362):k601
Cohen JC, Boerwinkle E, Mosley TH Jr, Hobbs HH (2006) Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med 354(12):1264–1272
Lopalco L (2010) CCR5: from natural resistance to a new anti-HIV strategy. Viruses 2(2):574–600
Swerdlow DI, Holmes MV, Kuchenbaecker KB, Engmann JE, Shah T, Sofat R et al (2012) The interleukin-6 receptor as a target for prevention of coronary heart disease: a Mendelian randomisation analysis. Lancet 379(9822):1214–1224
Gaziano L, Giambartolomei C, Pereira AC, Gaulton A, Posner DC, Swanson SA et al (2021) Actionable druggable genome-wide Mendelian randomization identifies repurposing opportunities for COVID-19. Nat Med 27(4):668–676
McCutcheon RA, Abi-Dargham A, Howes OD (2019) Schizophrenia, dopamine and the striatum: from biology to symptoms. Trends Neurosci 42(3):205–220
Abi-Dargham A (2014) Schizophrenia: overview and dopamine dysfunction. J Clin Psychiatry 75(11):e31
Grace AA (2016) Dysregulation of the dopamine system in the pathophysiology of schizophrenia and depression. Nat Rev Neurosci 17(8):524–532
Weinstein JJ, Chohan MO, Slifstein M, Kegeles LS, Moore H, Abi-Dargham A (2017) Pathway-specific dopamine abnormalities in schizophrenia. Biol Psychiatry 81(1):31–42
Jerber J, Seaton DD, Cuomo ASE, Kumasaka N, Haldane J, Steer J et al (2021) Population-scale single-cell RNA-seq profiling across dopaminergic neuron differentiation. Nat Genet 53(3):304–312
Kilpinen H, Goncalves A, Leha A, Afzal V, Alasoo K, Ashford S et al (2017) Common genetic variation drives molecular heterogeneity in human iPSCs. Nature 546(7658):370–375
Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D et al (2018) The MR-base platform supports systematic causal inference across the human phenome. Elife 30:7
Wang D, Liu S, Warrell J, Won H, Shi X, Navarro FCP et al (2018) Comprehensive functional genomic resource and integrative model for the human brain. Science (New York, NY) 362(6420):eaat8464
Nowakowski TJ, Bhaduri A, Pollen AA, Alvarado B, Mostajo-Radji MA, Di Lullo E et al (2017) Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Sci 358(6368):1318–1323
Speir ML, Bhaduri A, Markov NS, Moreno P, Nowakowski TJ, Papatheodorou I et al (2021) UCSC cell browser: visualize your single-cell data. Bioinforma 37(23):4578–4580
Li Y, Ling K, Hu J (2012) The emerging role of Arf/Arl small GTPases in cilia and ciliopathies. J Cell Biochem 113(7):2201–2207
Zhou C, Cunningham L, Marcus AI, Li Y, Kahn RA (2006) Arl2 and Arl3 regulate different microtubule-dependent processes. Mol Biol Cell 17(5):2476–2487
Jiang L, Rogers SL, Crews ST (2007) The Drosophila dead end Arf-like3 GTPase controls vesicle trafficking during tracheal fusion cell morphogenesis. Dev Biol 311(2):487–499
Yu H, Yan H, Li J, Li Z, Zhang X, Ma Y et al (2017) Common variants on 2p16.1, 6p22.1 and 10q24.32 are associated with schizophrenia in Han Chinese population. Mol Psychiatry 22(7):954–60
Liu J, Li X, Luo XJ (2021) Proteome-wide association study provides insights into the genetic component of protein abundance in psychiatric disorders. Biol Psychiatry 90(11):781–789
Ripke S, O’Dushlaine C, Chambert K, Moran JL, Kähler AK, Akterin S et al (2013) Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet 45(10):1150–1159
Mullins N, Forstner AJ, O’Connell KS, Coombes B, Coleman JRI, Qiao Z et al (2021) Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat Genet 53(6):817–829
van Hulzen KJE, Scholz CJ, Franke B, Ripke S, Klein M, McQuillin A et al (2017) Genetic overlap between attention-deficit/hyperactivity disorder and bipolar disorder: evidence from genome-wide association study meta-analysis. Biol Psychiatry 82(9):634–641
Nakashima A, Yamaguchi H, Kondo M, Furumura T, Kodani Y, Kaneko YS et al (2020) NT5DC2 affects the phosphorylation of tyrosine hydroxylase regulating its catalytic activity. J Neural Transm (Vienna) 127(12):1631–1640
Klein MO, Battagello DS, Cardoso AR, Hauser DN, Bittencourt JC, Correa RG (2019) Dopamine: functions, signaling, and association with neurological diseases. Cell Mol Neurobiol 39(1):31–59
Howes OD, Kapur S (2009) The dopamine hypothesis of schizophrenia: version III–the final common pathway. Schizophr Bull 35(3):549–562
Grace AA (2011) Dopamine system dysregulation by the hippocampus: implications for the pathophysiology and treatment of schizophrenia. Neuropharmacol 62(3):1342–1348
Lodge DJ, Grace AA (2011) Hippocampal dysregulation of dopamine system function and the pathophysiology of schizophrenia. Trends Pharmacol Sci 32(9):507–513
Purves-Tyson TD, Owens SJ, Rothmond DA, Halliday GM, Double KL, Stevens J et al (2017) Putative presynaptic dopamine dysregulation in schizophrenia is supported by molecular evidence from post-mortem human midbrain. Transl Psychiatry 7(1):e1003
Sonnenschein SF, Gomes FV, Grace AA (2020) Dysregulation of midbrain dopamine system and the pathophysiology of schizophrenia. Front Psychiatry 11:613
Hokfelt T, Ljungdahl A, Fuxe K, Johansson O (1974) Dopamine nerve terminals in the rat limbic cortex: aspects of the dopamine hypothesis of schizophrenia. Sci 184(4133):177–179
Luchins D (1975) The dopamine hypothesis of schizophrenia A critical analysis. Neuropsychobiology 1(6):365–378
Meltzer HY, Stahl SM (1976) The dopamine hypothesis of schizophrenia: a review. Schizophr Bull 2(1):19–76
Snyder SH (1976) The dopamine hypothesis of schizophrenia: focus on the dopamine receptor. Am J Psychiatry 133(2):197–202
McKenna PJ (1987) Pathology, phenomenology and the dopamine hypothesis of schizophrenia. Br J Psychiatry 151:288–301
Carlsson A (1988) The current status of the dopamine hypothesis of schizophrenia. Neuropsychopharmacol 1(3):179–186
Pilowsky LS, Costa DC, Ell PJ, Murray RM, Verhoeff NP, Kerwin RW (1992) Clozapine, single photon emission tomography, and the D2 dopamine receptor blockade hypothesis of schizophrenia. Lancet 340(8813):199–202
Toda M, Abi-Dargham A (2007) Dopamine hypothesis of schizophrenia: making sense of it all. Curr Psychiatry Rep 9(4):329–336
da Silva AF, Figee M, van Amelsvoort T, Veltman D, de Haan L (2008) The revised dopamine hypothesis of schizophrenia: evidence from pharmacological MRI studies with atypical antipsychotic medication. Psychopharmacol Bull 41(1):121–132
Deutch AY, Colbran RJ, Winder DJ (2007) Striatal plasticity and medium spiny neuron dendritic remodeling in parkinsonism. Parkinsonism Relat Disord 13(Suppl 3):S251–S258
Yang L, Gilbert ML, Zheng R, McKnight GS (2014) Selective expression of a dominant-negative type Ialpha PKA regulatory subunit in striatal medium spiny neurons impairs gene expression and leads to reduced feeding and locomotor activity. J Neurosci 34(14):4896–4904
Matoba N, Liang D, Sun H, Aygun N, McAfee JC, Davis JE et al (2020) Common genetic risk variants identified in the SPARK cohort support DDHD2 as a candidate risk gene for autism. Transl Psychiatry 10(1):265
Park CY, Zhou J, Wong AK, Chen KM, Theesfeld CL, Darnell RB et al (2021) Genome-wide landscape of RNA-binding protein target site dysregulation reveals a major impact on psychiatric disorder risk. Nat Genet 53(2):166–173
Hall LS, Pain O, O’Brien HE, Anney R, Walters JTR, Owen MJ et al (2021) Cis-effects on gene expression in the human prenatal brain associated with genetic risk for neuropsychiatric disorders. Mol Psychiatry 26(6):2082–2088
Ten Hagen KG, Fritz TA, Tabak LA (2003) All in the family: the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases. Glycobiology 13(1):1R-16R
Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J et al (2014) Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics 13(2):397–406
Perrine CL, Ganguli A, Wu P, Bertozzi CR, Fritz TA, Raman J et al (2009) Glycopeptide-preferring polypeptide GalNAc transferase 10 (ppGalNAc T10), involved in mucin-type O-glycosylation, has a unique GalNAc-O-Ser/Thr-binding site in its catalytic domain not found in ppGalNAc T1 or T2. J Biol Chem 284(30):20387–20397
Reily C, Stewart TJ, Renfrow MB, Novak J (2019) Glycosylation in health and disease. Nat Rev Nephrol 15(6):346–366
Freeze HH, Eklund EA, Ng BG, Patterson MC (2015) Neurological aspects of human glycosylation disorders. Annu Rev Neurosci 8(38):105–125
Williams SE, Mealer RG, Scolnick EM, Smoller JW, Cummings RD (2020) Aberrant glycosylation in schizophrenia: a review of 25 years of post-mortem brain studies. Mol Psychiatry 25(12):3198–3207
Schuurs-Hoeijmakers JH, Geraghty MT, Kamsteeg EJ, Ben-Salem S, de Bot ST, Nijhof B et al (2012) Mutations in DDHD2, encoding an intracellular phospholipase A(1), cause a recessive form of complex hereditary spastic paraplegia. Am J Hum Genet 91(6):1073–1081
Gonzalez M, Nampoothiri S, Kornblum C, Oteyza AC, Walter J, Konidari I et al (2013) Mutations in phospholipase DDHD2 cause autosomal recessive hereditary spastic paraplegia (SPG54). Eur J Hum Genet 21(11):1214–1218
Kumar KR, Blair NF, Sue CM (2015) An update on the hereditary spastic paraplegias: new genes and new disease models. Mov Disord Clin Pract 2(3):213–223
Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E et al (2006) Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutières syndrome and mimic congenital viral brain infection. Nat Genet 38(8):910–16
Rice G, Patrick T, Parmar R, Taylor CF, Aeby A, Aicardi J et al (2007) Clinical and molecular phenotype of Aicardi-Goutieres syndrome. Am J Hum Genet 81(4):713–725
Rice GI, Forte GM, Szynkiewicz M, Chase DS, Aeby A, Abdel-Hamid MS et al (2013) Assessment of interferon-related biomarkers in Aicardi-Goutières syndrome associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, and ADAR: a case-control study. Lancet Neurol 12(12):1159–1169
Vershinin Z, Feldman M, Werner T, Weil LE, Kublanovsky M, Abaev-Schneiderman E et al (2021) BRD4 methylation by the methyltransferase SETD6 regulates selective transcription to control mRNA translation. Sci Adv 7(22):eabf5374
Webb WM, Irwin AB, Pepin ME, Henderson BW, Huang V, Butler AA et al (2020) The SETD6 methyltransferase plays an essential role in hippocampus-dependent memory formation. Biol Psychiatry 87(6):577–587
Guo JY, Ragland JD, Carter CS (2019) Memory and cognition in schizophrenia. Mol Psychiatry 24(5):633–642
Acknowledgements
We thank miss Qian Li for her technical assistance.
Funding
This study was equally supported by the National Nature Science Foundation of China (U2102205 and 31970561 to XJL, 81830040 and 82130042 to ZJZ) and the Distinguished Young Scientists grant of the Yunnan Province (202001AV070006). This study was also supported by the Key Research Project of Yunnan Province (202101AS070055 to XJL), the Science and Technology of Guangdong province (2018B030334001 to ZJZ), the CAS “Light of West China” Program to JWL, and Yunnan Fundamental Research Projects (202001AT070099) to JWL.
Author information
Authors and Affiliations
Contributions
XJL conceived, designed, and supervised the whole study. XLD performed most of the analyses. JWL conducted the PPI analysis. XLD, JWL, XJL, and ZJZ interpreted the results. XJL and ZJZ wrote the manuscript. All authors provided critical comments and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval
Ethical approval is not required.
Consent to Participate
Not applicable.
Consent for Publication
The authors have given consent for publication.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dang, X., Liu, J., Zhang, Z. et al. Mendelian Randomization Study Using Dopaminergic Neuron-Specific eQTL Identifies Novel Risk Genes for Schizophrenia. Mol Neurobiol 60, 1537–1546 (2023). https://doi.org/10.1007/s12035-022-03160-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-022-03160-3