Abstract
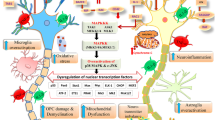
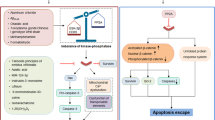
MYC is well known as a potent oncogene involved in regulating cell cycle and metabolism. Augmented MYC expression leads to cell cycle dysregulation, intense cell proliferation, and carcinogenesis. Surprisingly, its increased expression in neurons does not induce their proliferation, but leads to neuronal cell death and consequent development of a neurodegenerative phenotype. Interestingly, while cancer and neurodegenerative diseases such as Alzheimer’s disease are placed at the opposite sides of cell division spectrum, both start with cell cycle dysregulation and stimulation of proliferation. It seems that MYC action directed toward neuron cell proliferation and neural tissue repair collides with evolutional loss of regenerative capacity of CNS neurons in order to strengthen synaptic structure, to protect our cognitive abilities and therefore character. Accordingly, there are abundant mechanisms that block its expression and action specifically in the brain. Moreover, while MYC expression in brain neurons during neurodegenerative processes is related to their death, there are obvious evidences that MYC action after physical injury is beneficial in case of peripheral nerve recovery. MYC might be a useful tool to repair brain cells upon development of neurodegenerative disease or CNS trauma, including stroke and traumatic brain and spinal cord injury, as even imperfect axonal growth and regeneration strategies will likely be of profound benefit. Understanding complex control of MYC action in the brain might have important therapeutic significance, but also it may contribute to the comprehension of development of neurodegenerative diseases.


Similar content being viewed by others
References
Krafts KP (2010) Tissue repair: the hidden drama. Organogenesis 6:225–233. https://doi.org/10.4161/org.6.4.12555
Kalucka J, Missiaen R, Georgiadou M et al (2015) Metabolic control of the cell cycle. Cell Cycle 14:3379–3388. https://doi.org/10.1080/15384101.2015.1090068
Fritz V, Fajas L (2010) Metabolism and proliferation share common regulatory pathways in cancer cells. Oncogene 29:4369–4377. https://doi.org/10.1038/onc.2010.182
Conacci-Sorrell M, McFerrin L, Eisenman RN (2014) An overview of MYC and its interactome. Cold Spring Harb Perspect Med 4:1–24. https://doi.org/10.1101/cshperspect.a014357
Nesbit CE, Tersak JM, Prochownik EV (1999) MYC oncogenes and human neoplastic disease. Oncogene 18:3004–3016. https://doi.org/10.1038/sj.onc.1202746
Pelengaris S, Khan M, Evan G (2002) c-MYC: more than just a matter of life and death. Nat Rev Cancer 2:764–776. https://doi.org/10.1038/nrc904
Lancho O, Herranz D (2018) The MYC Enhancer-ome: long-range transcriptional regulation of MYC in cancer. Trends in Cancer 4:810–822. https://doi.org/10.1016/j.trecan.2018.10.003
Stine ZE, Walton ZE, Altman BJ et al (2015) MYC, metabolism, and cancer. Cancer Discov 5:1024–1039. https://doi.org/10.1158/2159-8290.CD-15-0507
Dang CV (2012) MYC on the path to cancer. Cell 149:22–35. https://doi.org/10.1016/j.cell.2012.03.003
Marinković D, Marinković T (2018) c-Myc misregulation triggers complex process of genomic instability. Genetika 50:731–745. https://doi.org/10.2298/GENSR1802731M
Marinkovic D, Marinkovic T, Mahr B et al (2004) Reversible lymphomagenesis in conditionally c-MYC expressing mice. Int J Cancer 110:336–342. https://doi.org/10.1002/ijc.20099
Felsher DW, Bishop JM (1999) Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell 4:199–207. https://doi.org/10.1016/S1097-2765(00)80367-6
Lee CVD, WM, (1999) c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol 19:1–11. https://doi.org/10.1128/mcb.19.1.1
Kokai E, Voss F, Fleischer F et al (2009) Myc regulates embryonic vascular permeability and remodeling. Circ Res 104:1151–1159. https://doi.org/10.1161/CIRCRESAHA.108.191460
Casey SC, Baylot V, Felsher DW (2018) The MYC oncogene is a global regulator of the immune response. Blood 131:2007–2015. https://doi.org/10.1182/blood-2017-11-742577
Marinkovic D, Marinkovic T (2020) The new role for an old guy: MYC as an immunoplayer. J Cell Physiol. https://doi.org/10.1002/jcp.30123
Kretzner L, Blackwood EM, Eisenman RN (1992) Myc and Max proteins possess distinct transcriptional activities. Nature 359:426–429. https://doi.org/10.1038/359426a0
Guo J, Li T, Schipper J et al (2014) Sequence specificity incompletely defines the genome-wide occupancy of Myc. Genome Biol 15:482. https://doi.org/10.1186/s13059-014-0482-3
Schneider A, Peukert K, Eilers M, Hänel F (1997) Association of Myc with the zinc-finger protein Miz-1 defines a novel pathway for gene regulation by Myc. Curr Top Microbiol Immunol 224:137–146. https://doi.org/10.1007/978-3-642-60801-8_14
Von Der Lehr N, Johansson S, Wu S et al (2003) The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell 11:1189–1200. https://doi.org/10.1016/S1097-2765(03)00193-X
Fernandez PC, Frank SR, Wang L et al (2003) Genomic targets of the human c-Myc protein. Genes Dev 17:1115–1129. https://doi.org/10.1101/gad.1067003
Rahl PB, Young RA (2014) MYC and transcription elongation. Cold Spring Harb Perspect Med 4:a020990. https://doi.org/10.1101/cshperspect.a020990
Zhang Y, Liu T, Meyer CA et al (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9:R137. https://doi.org/10.1186/gb-2008-9-9-r137
Nie Z, Hu G, Wei G et al (2012) c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151:68–79. https://doi.org/10.1016/j.cell.2012.08.033
Lin CY, Lovén J, Rahl PB et al (2012) Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151:56–67. https://doi.org/10.1016/j.cell.2012.08.026
Dang CV, O’Donnell KA, Zeller KI et al (2006) The c-Myc target gene network. Semin Cancer Biol 16:253–264. https://doi.org/10.1016/j.semcancer.2006.07.014
Shchors K, Shchors E, Rostker F et al (2006) The Myc-dependent angiogenic switch in tumors is mediated by interleukin 1β. Genes Dev 20:2527–2538. https://doi.org/10.1101/gad.1455706
Kortlever RM, Sodir NM, Wilson CH et al (2017) Myc cooperates with Ras by programming inflammation and immune suppression. Cell 171:1301-1315.e14. https://doi.org/10.1016/j.cell.2017.11.013
Bangert A, Cristofanon S, Eckhardt I et al (2012) Histone deacetylase inhibitors sensitize glioblastoma cells to TRAIL-induced apoptosis by c-myc-mediated downregulation of cFLIP. Oncogene 31:4677–4688. https://doi.org/10.1038/onc.2011.614
Vjetrovic J, Shankaranarayanan P, Mendoza-Parra MA, Gronemeyer H (2014) Senescence-secreted factors activate Myc and sensitize pretransformed cells to TRAIL-induced apoptosis. Aging Cell 13:487–496. https://doi.org/10.1111/acel.12197
Bywater MJ, Burkhart DL, Straube J et al (2020) Reactivation of Myc transcription in the mouse heart unlocks its proliferative capacity. Nat Commun 11:1827. https://doi.org/10.1038/s41467-020-15552-x
Lee HG, Casadesus G, Nunomura A et al (2009) The neuronal expression of MYC causes a neurodegenerative phenotype in a novel transgenic mouse. Am J Pathol 174:891–897. https://doi.org/10.2353/ajpath.2009.080583
Lee H, gon, Casadesus G, Zhu X, et al (2009) Cell cycle re-entry mediated neurodegeneration and its treatment role in the pathogenesis of Alzheimer’s disease. Neurochem Int 54:84–88. https://doi.org/10.1016/j.neuint.2008.10.013
Nagy Z, Esiri MM, Smith AD (1997) Expression of cell division markers in the hippocampus in Alzheimer’s disease and other nenrodegenerative conditions. Acta Neuropathol 93:294–300. https://doi.org/10.1007/s004010050617
Zhu X, McShea A, Harris PLR et al (2004) Elevated expression of a regulator of the G2/M phase of the cell cycle, neuronal CIP-1-associated regulator of cyclin B, in Alzheimer’s disease. J Neurosci Res 75:698–703. https://doi.org/10.1002/jnr.20028
Mosch B, Morawski M, Mittag A et al (2007) Aneuploidy and DNA replication in the normal human brain and Alzheimer’s disease. J Neurosci 27:6859–6867. https://doi.org/10.1523/JNEUROSCI.0379-07.2007
Yang Y, Geldmacher DS, Herrup K (2001) DNA replication precedes neuronal cell death in Alzheimer’s disease. J Neurosci 21:2661–2668. https://doi.org/10.1523/jneurosci.21-08-02661.2001
Majd S, Zarifkar A, Rastegar K, Takhshid MA (2008) Different fibrillar Aβ 1–42 concentrations induce adult hippocampal neurons to reenter various phases of the cell cycle. Brain Res 1218:224–229. https://doi.org/10.1016/j.brainres.2008.04.050
Woods J, Snape M, Smith MA (2007) The cell cycle hypothesis of Alzheimer’s disease: suggestions for drug development. Biochim Biophys Acta - Mol Basis Dis 1772:503–508. https://doi.org/10.1016/j.bbadis.2006.12.004
Snape M, Lee HG, Casadesus G, Smith MA (2009) Cell cycle aberrations in Alzheimer’s disease: a novel therapeutic opportunity. Expert Rev Neurother 9:1579–1580. https://doi.org/10.1586/ern.09.113
Lopes J, Oliveira C, Agostinho P (2009) Cell cycle re-entry in alzheimers disease: a major neuropathological characteristic? Curr Alzheimer Res 6:205–212. https://doi.org/10.2174/156720509788486590
Jordan-Sciutto KL, Dorsey R, Chalovich EM et al (2003) Expression patterns of retinoblastoma protein in Parkinson disease. J Neuropathol Exp Neurol 62:68–74. https://doi.org/10.1093/jnen/62.1.68
Ranganathan S, Bowser R (2003) Alterations in G1 to S phase cell-cycle regulators during amyotrophic lateral sclerosis. Am J Pathol 162:823–835. https://doi.org/10.1016/S0002-9440(10)63879-5
Ferrer I, Blanco R, Carmona M, Puig B (2001) Phosphorylated c-MYC expression in Alzheimer disease, Pick’s disease, progressive supranuclear palsy and corticobasal degeneration. Neuropathol Appl Neurobiol 27:343–351. https://doi.org/10.1046/j.1365-2990.2001.00348.x
Ferrer I, Blanco R (2000) N-myc and c-myc expression in Alzheimer disease, Huntington disease and Parkinson disease. Mol Brain Res 77:270–276. https://doi.org/10.1016/S0169-328X(00)00062-0
Lee HP, Kudo W, Zhu X et al (2011) Early induction of c-Myc is associated with neuronal cell death. Neurosci Lett 505:124–127. https://doi.org/10.1016/j.neulet.2011.10.004
Sleiman SF, Langley BC, Basso M et al (2011) Mithramycin is a gene-selective sp1 inhibitor that identifies a biological intersection between cancer and neurodegeneration. J Neurosci 31:6858–6870. https://doi.org/10.1523/JNEUROSCI.0710-11.2011
Dhillon AS, Hagan S, Rath O, Kolch W (2007) MAP kinase signalling pathways in cancer. Oncogene 26:3279–3290. https://doi.org/10.1038/sj.onc.1210421
Marshall GM, Liu PY, Gherardi S et al (2011) SIRT1 promotes N-Myc oncogenesis through a positive feedback loop involving the effects of MKP3 and ERK on N-Myc protein stability. PLoS Genet 7:e1002135. https://doi.org/10.1371/journal.pgen.1002135
Porta C, Paglino C, Mosca A (2014) Targeting PI3K/Akt/mTOR signaling in cancer. Front Oncol 4:64. https://doi.org/10.3389/fonc.2014.00064
Pei JJ, Hugon J (2008) mTOR-dependent signalling in Alzheimer’s disease. J Cell Mol Med 12:2525–2532
White MC, Holman DM, Boehm JE et al (2014) Age and cancer risk: a potentially modifiable relationship. Am J Prev Med 46:S7-15. https://doi.org/10.1016/j.amepre.2013.10.029
Guerreiro R, Bras J (2015) The age factor in Alzheimer’s disease. Genome Med 7:1–3. https://doi.org/10.1186/s13073-015-0232-5
Musicco M, Adorni F, Di Santo S et al (2013) Inverse occurrence of cancer and Alzheimer disease: a population-based incidence study. Neurology 81:322–328. https://doi.org/10.1212/WNL.0b013e31829c5ec1
Majd S, Power J, Majd Z (2019) Alzheimer’s Disease and cancer: when two monsters cannot be together. Front Neurosci 13:155. https://doi.org/10.3389/fnins.2019.00155
Hay N (2011) Interplay between FOXO, TOR, and Akt. Biochim Biophys Acta - Mol Cell Res 1813:1965–1970. https://doi.org/10.1016/j.bbamcr.2011.03.013
Gómez-Crisóstomo NP, Rodríguez Martínez E, Rivas-Arancibia S (2014) Oxidative stress activates the transcription factors FoxO 1a and FoxO 3a in the hippocampus of rats exposed to low doses of ozone. Oxid Med Cell Longev 2014:805764. https://doi.org/10.1155/2014/805764
Bullock M (2016) FOXO factors and breast cancer: outfoxing endocrine resistance. Endocr Relat Cancer 23:R113–R130. https://doi.org/10.1530/ERC-15-0461
Jiramongkol Y, Lam EWF (2020) FOXO transcription factor family in cancer and metastasis. Cancer Metastasis Rev 39:681–709. https://doi.org/10.1007/s10555-020-09883-w
Chandramohan V, Jeay S, Pianetti S, Sonenshein GE (2004) Reciprocal control of forkhead box O 3a and c-Myc via the phosphatidylinositol 3-kinase pathway coordinately regulates p27 Kip1 levels. J Immunol 172:5522–5527. https://doi.org/10.4049/jimmunol.172.9.5522
Riddell M, Nakayama A, Hikita T et al (2018) aPKC controls endothelial growth by modulating c-Myc via FoxO1 DNA-binding ability. Nat Commun 9:5357. https://doi.org/10.1038/s41467-018-07739-0
Ariga H (2015) Common mechanisms of onset of cancer and neurodegenerative diseases. Biol Pharm Bull 38:795–808. https://doi.org/10.1248/bpb.b15-00125
Qin ZH, Chen RW, Wang Y et al (1999) Nuclear factor κB nuclear translocation upregulates c-Myc and p53 expression during NMDA receptor-mediated apoptosis in rat striatum. J Neurosci 19:4023–4033. https://doi.org/10.1523/jneurosci.19-10-04023.1999
Arendt T (2003) Synaptic plasticity and cell cycle activation in neurons are alternative effector pathways: the “Dr. Jekyll and Mr. Hyde concept” of Alzheimer’s disease or the yin and yang of neuroplasticity. Prog Neurobiol 71:83–248. https://doi.org/10.1016/j.pneurobio.2003.09.007
Klein JA, Ackerman SL (2003) Oxidative stress, cell cycle, and neurodegeneration. J Clin Invest 111:785–793. https://doi.org/10.1172/jci18182
Sheen J-H, Dickson RB (2002) Overexpression of c-Myc alters G1/S arrest following ionizing radiation. Mol Cell Biol 22:1819–1833. https://doi.org/10.1128/mcb.22.6.1819-1833.2002
Li Q, Dang CV (1999) c-Myc overexpression uncouples DNA replication from mitosis. Mol Cell Biol 19:5339–5351. https://doi.org/10.1128/mcb.19.8.5339
McGahan L, Hakim AM, Robertson GS (1998) Hippocampal Myc and p53 expression following transient global ischemia. Mol Brain Res 56:133–145. https://doi.org/10.1016/S0169-328X(98)00038-2
Purves D, Augustine GJ, Fitzpatrick D, Katz LC, LaMantia A-S, McNamaraWilliams JOSM (2001) Neuroscience, 2nd edn. Sinauer Associates, Sunderland
Belin S, Nawabi H, Wang C et al (2015) Injury-induced decline of intrinsic regenerative ability revealed by quantitative proteomics. Neuron 86:1000–1014. https://doi.org/10.1016/j.neuron.2015.03.060
Ma JJ, Ju X, Xu RJ et al (2019) Telomerase reverse transcriptase and p53 regulate mammalian peripheral nervous system and CNS axon regeneration downstream of c-Myc. J Neurosci 39:9107–9118. https://doi.org/10.1523/JNEUROSCI.0419-19.2019
Maciejowski J, De Lange T (2017) Telomeres in cancer: tumour suppression and genome instability. Nat Rev Mol Cell Biol 18:175–186. https://doi.org/10.1038/nrm.2016.171
Holtmaat A, Svoboda K (2009) Experience-dependent structural synaptic plasticity in the mammalian brain. Nat Rev Neurosci 10:647–658. https://doi.org/10.1038/nrn2699
Wang Y, Liu Y, Chen Y et al (2009) Peripheral nerve injury induces down-regulation of Foxo3a and p27 kip1 in rat dorsal root ganglia. Neurochem Res 34:891–898. https://doi.org/10.1007/s11064-008-9849-8
Hasmatali JD, De Guzman J, Johnston J et al (2020) FOXO3a as a sensor of unilateral nerve injury in sensory neurons ipsilateral, contralateral and remote to injury. Neural Regen Res 15:2353. https://doi.org/10.4103/1673-5374.284999
Groves MJ, Schänzer A, Simpson AJ et al (2003) Profile of adult rat sensory neuron loss, apoptosis and replacement after sciatic nerve crush. J Neurocytol 32:113–122. https://doi.org/10.1023/B:NEUR.0000005596.88385.ec
Xian CJ, Zhou XF (1999) Neuronal-glial differential expression of TGF-α and its receptor in the dorsal root ganglia in response to sciatic nerve lesion. Exp Neurol 157:317–326. https://doi.org/10.1006/exnr.1999.7063
Santo EE, Paik J (2018) FOXO in neural cells and diseases of the nervous system. Curr Top Dev Biol 127:105–118. https://doi.org/10.1016/bs.ctdb.2017.10.002
Sun F, Park KK, Belin S et al (2011) Sustained axon regeneration induced by co-deletion of PTEN and SOCS3. Nature 480:372–375. https://doi.org/10.1038/nature10594
Tanaka EM, Ferretti P (2009) Considering the evolution of regeneration in the central nervous system. Nat Rev Neurosci 10:713–723. https://doi.org/10.1038/nrn2707
Singh MD, Raj K, Sarkar S (2014) Drosophila Myc, a novel modifier suppresses the poly(Q) toxicity by modulating the level of CREB binding protein and histone acetylation. Neurobiol Dis 63:48–61. https://doi.org/10.1016/j.nbd.2013.11.015
Nandakumar S, Grushko O, Buttitta LA (2020) Polyploidy in the adult drosophila brain. Elife 9:1–25. https://doi.org/10.7554/ELIFE.54385
Tedeschi A, Omura T, Costigan M (2017) CNS repair and axon regeneration: using genetic variation to determine mechanisms. Exp Neurol 287:409–422. https://doi.org/10.1016/j.expneurol.2016.05.004
Funding
This work was financially supported by the Serbian Ministry of Education, Science and Technological Research (Grant No. 175083).
Author information
Authors and Affiliations
Contributions
The idea for this article came from Dragan Marinkovic, while both authors contributed in literature search and data analysis, structuring, drafting, writing and critical revising.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
Not applicable for the review paper.
Consent for Publication
Not applicable for the review paper.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Marinkovic, T., Marinkovic, D. Obscure Involvement of MYC in Neurodegenerative Diseases and Neuronal Repair. Mol Neurobiol 58, 4169–4177 (2021). https://doi.org/10.1007/s12035-021-02406-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-021-02406-w