Abstract
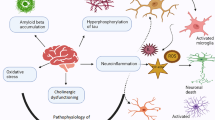
Lafora disease (LD) is a rare, fatal form of progressive myoclonus epilepsy. The molecular basis of this devastating disease is still poorly understood, and no treatment is available yet, which leads to the death of the patients around 10 years from the onset of the first symptoms. The hallmark of LD is the accumulation of insoluble glycogen-like inclusions in the brain and peripheral tissues, as a consequence of altered glycogen homeostasis. In addition, other determinants in the pathophysiology of LD have been suggested, such as proteostasis impairment, with reduction in autophagy, and oxidative stress, among others. In order to gain a general view of the genes involved in the pathophysiology of LD, in this work, we have performed RNA-Seq transcriptome analyses of whole-brain tissue from two independent mouse models of the disease, namely Epm2a−/− and Epm2b−/− mice, at different times of age. Our results provide strong evidence for three major facts: first, in both models of LD, we found a common set of upregulated genes, most of them encoding mediators of inflammatory response; second, there was a progression with the age in the appearance of these inflammatory markers, starting at 3 months of age; and third, reactive glia was responsible for the expression of these inflammatory genes. These results clearly indicate that neuroinflammation is one of the most important traits to be considered in order to fully understand the pathophysiology of LD, and define reactive glia as novel therapeutic targets in the disease.







Similar content being viewed by others
Abbreviations
- FC:
-
Fold change
- FDR:
-
False discovery rate
- LD:
-
Lafora disease
- RT-qPCR:
-
Real-time quantitative polymerase chain reaction
References
Turnbull J, Tiberia E, Striano P, Genton P, Carpenter S, Ackerley CA, Minassian BA (2016) Lafora disease. Epileptic Disord 18(S2):38–62
Garcia-Gimeno MA, Knecht E, Sanz P (2018) Lafora disease: a ubiquitination-related pathology. Cells 7(8)
Minassian BA, Lee JR, Herbrick JA, Huizenga J, Soder S, Mungall AJ, Dunham I, Gardner R et al (1998) Mutations in a gene encoding a novel protein tyrosine phosphatase cause progressive myoclonus epilepsy. Nat Genet 20(2):171–174
Serratosa JM, Gomez-Garre P, Gallardo ME, Anta B, de Bernabe DB, Lindhout D, Augustijn PB, Tassinari CA et al (1999) A novel protein tyrosine phosphatase gene is mutated in progressive myoclonus epilepsy of the Lafora type (EPM2). Hum Mol Genet 8(2):345–352
Chan EM, Young EJ, Ianzano L, Munteanu I, Zhao X, Christopoulos CC, Avanzini G, Elia M et al (2003) Mutations in NHLRC1 cause progressive myoclonus epilepsy. Nat Genet 35(2):125–127
Gentry MS, Worby CA, Dixon JE (2005) Insights into Lafora disease: malin is an E3 ubiquitin ligase that ubiquitinates and promotes the degradation of laforin. Proc Natl Acad Sci U S A 102(24):8501–8506
Lohi H, Ianzano L, Zhao XC, Chan EM, Turnbull J, Scherer SW, Ackerley CA, Minassian BA (2005) Novel glycogen synthase kinase 3 and ubiquitination pathways in progressive myoclonus epilepsy. Hum Mol Genet 14(18):2727–2736
Solaz-Fuster MC, Gimeno-Alcaniz JV, Ros S, Fernandez-Sanchez ME, Garcia-Fojeda B, Criado Garcia O, Vilchez D, Dominguez J et al (2008) Regulation of glycogen synthesis by the laforin-malin complex is modulated by the AMP-activated protein kinase pathway. Hum Mol Genet 17(5):667–678
Roma-Mateo C, Aguado C, Garcia-Gimenez JL, Knecht E, Sanz P, Pallardo FV (2015) Oxidative stress, a new hallmark in the pathophysiology of Lafora progressive myoclonus epilepsy. Free Radic Biol Med 88(Pt A):30–41
Gómez-Abad C, Gómez-Garre P, Gutiérrez-Delicado E, Saygi S, Michelucci R, Tassinari CA, Rodriguez de Cordoba S, Serratosa JM (2005) Lafora disease due to EPM2B mutations. A clinical and genetic study. Neurology 64:982–986
Ganesh S, Delgado-Escueta AV, Sakamoto T, Avila MR, Machado-Salas J, Hoshii Y, Akagi T, Gomi H et al (2002) Targeted disruption of the Epm2a gene causes formation of Lafora inclusion bodies, neurodegeneration, ataxia, myoclonus epilepsy and impaired behavioral response in mice. Hum Mol Genet 11(11):1251–1262
DePaoli-Roach AA, Tagliabracci VS, Segvich DM, Meyer CM, Irimia JM, Roach PJ (2010) Genetic depletion of the malin E3 ubiquitin ligase in mice leads to Lafora bodies and the accumulation of insoluble laforin. J Biol Chem 285(33):25372–25381
Valles-Ortega J, Duran J, Garcia-Rocha M, Bosch C, Saez I, Pujadas L, Serafin A, Canas X et al (2011) Neurodegeneration and functional impairments associated with glycogen synthase accumulation in a mouse model of Lafora disease. EMBO Molecular Medicine 3(11):667–681
Criado O, Aguado C, Gayarre J, Duran-Trio L, Garcia-Cabrero AM, Vernia S, San Millan B, Heredia M et al (2012) Lafora bodies and neurological defects in malin-deficient mice correlate with impaired autophagy. Hum Mol Genet 21(7):1521–1533
Garcia-Cabrero AM, Marinas A, Guerrero R, de Cordoba SR, Serratosa JM, Sanchez MP (2012) Laforin and malin deletions in mice produce similar neurologic impairments. J Neuropathol Exp Neurol 71(5):413–421
Garcia-Cabrero AM, Sanchez-Elexpuru G, Serratosa JM, Sanchez MP (2014) Enhanced sensitivity of laforin- and malin-deficient mice to the convulsant agent pentylenetetrazole. Front Neurosci 8:291
Aguado C, Sarkar S, Korolchuk VI, Criado O, Vernia S, Boya P, Sanz P, de Cordoba SR et al (2010) Laforin, the most common protein mutated in Lafora disease, regulates autophagy. Hum Mol Genet 19(14):2867–2876
Puri R, Ganesh S (2010) Laforin in autophagy: a possible link between carbohydrate and protein in Lafora disease? Autophagy 6(8):1229–1231
Turnbull J, DePaoli-Roach AA, Zhao X, Cortez MA, Pencea N, Tiberia E, Piliguian M, Roach PJ et al (2011) PTG depletion removes Lafora bodies and rescues the fatal epilepsy of Lafora disease. PLoS Genet 7(4):e1002037
Lopez-Gonzalez I, Viana R, Sanz P, Ferrer I (2017) Inflammation in Lafora disease: evolution with disease progression in laforin and malin knock-out mouse models. Mol Neurobiol 54(5):3119–3130
Rai A, Mishra R, Ganesh S (2017) Suppression of leptin signaling reduces polyglucosan inclusions and seizure susceptibility in a mouse model for Lafora disease. Hum Mol Genet 26(24):4778–4785
Bottomly D, Walter NA, Hunter JE, Darakjian P, Kawane S, Buck KJ, Searles RP, Mooney M et al (2011) Evaluating gene expression in C57BL/6J and DBA/2J mouse striatum using RNA-Seq and microarrays. PLoS One 6(3):e17820
Wang C, Gong B, Bushel PR, Thierry-Mieg J, Thierry-Mieg D, Xu J, Fang H, Hong H et al (2014) The concordance between RNA-seq and microarray data depends on chemical treatment and transcript abundance. Nat Biotechnol 32(9):926–932
Robinson MD, Oshlack A (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11(3):R25
Robinson MD, Smyth GK (2007) Moderated statistical tests for assessing differences in tag abundance. Bioinformatics 23(21):2881–2887
Robinson MD, Smyth GK (2008) Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 9(2):321–332
Lun AT, Chen Y, Smyth GK (2016) It’s DE-licious: a recipe for differential expression analyses of RNA-seq experiments using quasi-likelihood methods in edgeR. Methods Mol Biol 1418:391–416
Benjamini YaH Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B 57:289–300
da Huang W, Sherman BT, Stephens R, Baseler MW, Lane HC, Lempicki RA (2008) DAVID gene ID conversion tool. Bioinformation 2(10):428–430
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res 40(15):e115
Ganesh S, Tsurutani N, Amano K, Mittal S, Uchikawa C, Delgado-Escueta AV, Yamakawa K (2005) Transcriptional profiling of a mouse model for Lafora disease reveals dysregulation of genes involved in the expression and modification of proteins. Neurosci Lett 387(2):62–67
Holtman IR, Raj DD, Miller JA, Schaafsma W, Yin Z, Brouwer N, Wes PD, Moller T et al (2015) Induction of a common microglia gene expression signature by aging and neurodegenerative conditions: a co-expression meta-analysis. Acta Neuropathol Commun 3:31
Kang SS, Ren Y, Liu CC, Kurti A, Baker KE, Bu G, Asmann Y, Fryer JD (2018) Lipocalin-2 protects the brain during inflammatory conditions. Mol Psychiatry 23(2):344–350
Srinivasan K, Friedman BA, Larson JL, Lauffer BE, Goldstein LD, Appling LL, Borneo J, Poon C et al (2016) Untangling the brain’s neuroinflammatory and neurodegenerative transcriptional responses. Nat Commun 7:11295
Zamanian JL, Xu L, Foo LC, Nouri N, Zhou L, Giffard RG, Barres BA (2012) Genomic analysis of reactive astrogliosis. J Neurosci 32(18):6391–6410
Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, Phatnani HP, Guarnieri P et al (2014) An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34(36):11929–11947
Bosco DB, Zheng J, Xu Z, Peng J, Eyo UB, Tang K, Yan C, Huang J et al (2018) RNAseq analysis of hippocampal microglia after kainic acid-induced seizures. Mol Brain 11(1):34
Vezzani A, Balosso S, Ravizza T (2019) Neuroinflammatory pathways as treatment targets and biomarkers in epilepsy. Nat Rev Neurol 15(8):459–472
Tegelberg S, Kopra O, Joensuu T, Cooper JD, Lehesjoki AE (2012) Early microglial activation precedes neuronal loss in the brain of the Cstb−/− mouse model of progressive myoclonus epilepsy, EPM1. J Neuropathol Exp Neurol 71(1):40–53
Joensuu T, Tegelberg S, Reinmaa E, Segerstrale M, Hakala P, Pehkonen H, Korpi ER, Tyynela J et al (2014) Gene expression alterations in the cerebellum and granule neurons of Cstb(−/−) mouse are associated with early synaptic changes and inflammation. PLoS One 9(2):e89321
Okuneva O, Korber I, Li Z, Tian L, Joensuu T, Kopra O, Lehesjoki AE (2015) Abnormal microglial activation in the Cstb(−/−) mouse, a model for progressive myoclonus epilepsy, EPM1. Glia 63(3):400–411
Okuneva O, Li Z, Korber I, Tegelberg S, Joensuu T, Tian L, Lehesjoki AE (2016) Brain inflammation is accompanied by peripheral inflammation in Cstb (−/−) mice, a model for progressive myoclonus epilepsy. J Neuroinflammation 13(1):298
Rubio-Villena C, Viana R, Bonet J, Garcia-Gimeno MA, Casado M, Heredia M, Sanz P (2018) Astrocytes: new players in progressive myoclonus epilepsy of Lafora type. Hum Mol Genet 27(7):1290–1300
Matin N, Tabatabaie O, Falsaperla R, Lubrano R, Pavone P, Mahmood F, Gullotta M, Serra A et al (2015) Epilepsy and innate immune system: a possible immunogenic predisposition and related therapeutic implications. Hum Vaccin Immunother 11(8):2021–2029
Aronica E, Bauer S, Bozzi Y, Caleo M, Dingledine R, Gorter JA, Henshall DC, Kaufer D et al (2017) Neuroinflammatory targets and treatments for epilepsy validated in experimental models. Epilepsia 58(Suppl 3):27–38
Acknowledgments
We want to thank Miguel Heredia (CIBERER) for his help in obtaining the brain samples and Ursula Estada (Multigenic Analysis Unit from the UCIM-INCLIVA; University of Valencia, Valencia, Spain) for her help in constructing libraries and performing RNA-Seq experiments.
Funding
This work was supported by grants from the Spanish Ministry of Economy and Competitiveness SAF2014-54604-C3-1-R and SAF2017-83151-R, a grant from Fundación Ramón Areces (CIVP18A3935), and a grant from the National Institute of Health (NIH-NINDS) P01NS097197, which established the Lafora Epilepsy Cure Initiative (LECI), to PS. We also acknowledge a grant from the Spanish Ministry of Economy and Competitiveness SAF2014-54604-C3-2-R to EK and a grant from Generalitat Valenciana Prometeo2018/135 to PS and FVP.
Author information
Authors and Affiliations
Contributions
ML, CA, and AF prepared the samples for RNA-Seq and performed the RT-qPCR; ML and MM-E conducted the western blot analyses; MM-E carried out the immunofluorescence analyses; DG and JLG-G performed the bioinformatics analyses; ML, MM-E, and CR-M analyzed the data; and EK, FVP, and PS interpreted the data and wrote the manuscript.
Corresponding author
Ethics declarations
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Consejo Superior de Investigaciones Cientificas (CSIC, Spain) and approved by the Consellería de Agricultura, Medio Ambiente, Cambio Climático y Desarrollo Rural from the Generalitat Valenciana. All mouse procedures were approved by the animal committee of the Instituto de Biomedicina de Valencia-CSIC [Permit Number: (IBV-16), 2015/VSC/PEA/00029].
Competing Interests
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Marcos Lahuerta and Daymé Gonzalez should be regarded as joint first authors.
Electronic Supplementary Material
ESM 1
(DOCX 775 kb)
Rights and permissions
About this article
Cite this article
Lahuerta, M., Gonzalez, D., Aguado, C. et al. Reactive Glia-Derived Neuroinflammation: a Novel Hallmark in Lafora Progressive Myoclonus Epilepsy That Progresses with Age. Mol Neurobiol 57, 1607–1621 (2020). https://doi.org/10.1007/s12035-019-01842-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-019-01842-z