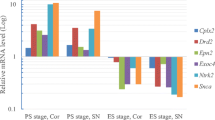
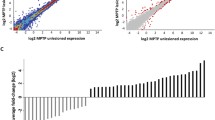
Abstract
Parkinson’s disease (PD) is characterized by degeneration of dopaminergic neurons. A whole-transcriptome analysis of the substantia nigra and striatum of an MPTP-induced mouse models of the earliest stages of PD was performed. Functional clustering of differentially represented transcripts revealed processes associated with the functioning of synapses, dendrites, axons, and myelination of neuronal projections. All of these processes occur in both the substantia nigra and striatum, but they are aimed at the functioning of neuron terminals in the striatum. One cluster was identified at the earliest stage modeled, i.e., “neuron projection” in the substantia nigra and “transport” in the striatum, and their number increased at subsequent stages. The number of clusters in the striatum predominates over those in the substantia nigra and there is a pronounced increase in the number of clusters from the modeled early stages to the late stages. These findings indicate that the substantia nigra and striatum have unique patterns of changes at each stage. Considering the clustering of individual processes, it was seen that there is a set of hierarchical clusters that overlap only partially at different stages and in different tissues. The data indicate a consistent involvement of the transcriptome in the pathogenesis of PD and highlight the independent role of various brain structures and individual parts of nerve cells in the formation of a response to the development of neurodegeneration. Decreased myelination of neuronal projections may be associated with the development of PD in the models considered.





Similar content being viewed by others
References
Racette BA, Tabbal SD, Jennings D, Good L, Perlmutter JS, Evanoff B (2005) Prevalence of parkinsonism and relationship to exposure in a large sample of Alabama welders. Neurology 64(2):230–235. https://doi.org/10.1212/01.WNL.0000149511.19487.44
Claveria LE, Duarte J, Sevillano MD, Perez-Sempere A, Cabezas C, Rodriguez F, de Pedro-Cuesta J (2002) Prevalence of Parkinson’s disease in Cantalejo, Spain: a door-to-door survey. Mov Disord 17(2):242–249. https://doi.org/10.1002/mds.10087
van de Vijver DA, Roos RA, Jansen PA, Porsius AJ, de Boer A (2001) Estimation of incidence and prevalence of Parkinson’s disease in the elderly using pharmacy records. Pharmacoepidemiol Drug Saf 10(6):549–554. https://doi.org/10.1002/pds.624
Chan DK, Cordato D, Karr M, Ong B, Lei H, Liu J, Hung WT (2005) Prevalence of Parkinson’s disease in Sydney. Acta Neurol Scand 111(1):7–11. https://doi.org/10.1111/j.1600-0404.2004.00348.x
Morioka S, Sakata K, Yoshida S, Nakai E, Shiba M, Yoshimura N, Hashimoto T (2002) Incidence of Parkinson disease in Wakayama, Japan. J Epidemiol/Japan Epidemiol Assoc 12(6):403–407. https://doi.org/10.2188/jea.12.403
Bernheimer H, Birkmayer W, Hornykiewicz O, Jellinger K, Seitelberger F (1973) Brain dopamine and the syndromes of Parkinson and Huntington. Clinical, morphological and neurochemical correlations. J Neurol Sci 20(4):415–455. https://doi.org/10.1016/0022-510X(73)90175-5
Cookson MR, Hardy J, Lewis PA (2008) Genetic neuropathology of Parkinson’s disease. Int J Clin Exp Pathol 1(3):217–231
Fowler CJ (2007) Update on the neurology of Parkinson’s disease. Neurourol Urodyn 26(1):103–109. https://doi.org/10.1002/nau.20371
Hunn BH, Cragg SJ, Bolam JP, Spillantini MG, Wade-Martins R (2015) Impaired intracellular trafficking defines early Parkinson’s disease. Trends Neurosci 38(3):178–188. https://doi.org/10.1016/j.tins.2014.12.009
Trinh J, Farrer M (2013) Advances in the genetics of Parkinson disease. Nat Rev Neurol 9(8):445–454. https://doi.org/10.1038/nrneurol.2013.132
Lesage S, Brice A (2012) Role of Mendelian genes in “sporadic” Parkinson’s disease. Parkinsonism Relat Disord 18(Suppl 1):S66–S70. https://doi.org/10.1016/S1353-8020(11)70022-0
Langston JW, Ballard P, Tetrud JW, Irwin I (1983) Chronic Parkinsonism in humans due to a product of meperidine-analog synthesis. Science (New York, NY) 219(4587):979–980. https://doi.org/10.1126/science.6823561
German DC, Nelson EL, Liang CL, Speciale SG, Sinton CM, Sonsalla PK (1996) The neurotoxin MPTP causes degeneration of specific nucleus A8, A9 and A10 dopaminergic neurons in the mouse. Neurodegeneration 5(4):299–312. https://doi.org/10.1006/neur.1996.0041
Meredith GE, Totterdell S, Potashkin JA, Surmeier DJ (2008) Modeling PD pathogenesis in mice: advantages of a chronic MPTP protocol. Parkinsonism Relat Disord 14(Suppl 2):S112–S115. https://doi.org/10.1016/j.parkreldis.2008.04.012
Langston JW, Forno LS, Tetrud J, Reeves AG, Kaplan JA, Karluk D (1999) Evidence of active nerve cell degeneration in the substantia nigra of humans years after 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine exposure. Ann Neurol 46(4):598–605. https://doi.org/10.1002/1531-8249(199910)46:4<598::AID-ANA7>3.0.CO;2-F
Smeyne RJ, Jackson-Lewis V (2005) The MPTP model of Parkinson’s disease. Brain Res Mol Brain Res 134(1):57–66. https://doi.org/10.1016/j.molbrainres.2004.09.017
Prediger RD, Aguiar AS Jr, Rojas-Mayorquin AE, Figueiredo CP, Matheus FC, Ginestet L, Chevarin C, Bel ED et al (2010) Single intranasal administration of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine in C57BL/6 mice models early preclinical phase of Parkinson’s disease. Neurotox Res 17(2):114–129. https://doi.org/10.1007/s12640-009-9087-0
Dovero S, Gross C, Bezard E (2016) Unexpected toxicity of very low dose MPTP in mice: a clue to the etiology of Parkinson’s disease? Synapse (New York, NY) 70(2):49–51. https://doi.org/10.1002/syn.21875
Ugrumov MV, Khaindrava VG, Kozina EA, Kucheryanu VG, Bocharov EV, Kryzhanovsky GN, Kudrin VS, Narkevich VB et al (2011) Modeling of presymptomatic and symptomatic stages of parkinsonism in mice. Neuroscience 181:175–188. https://doi.org/10.1016/j.neuroscience.2011.03.007
Kolacheva AA, Kozina EA, Volina EV, Ugryumov MV (2014) Time course of degeneration of dopaminergic neurons and respective compensatory processes in the nigrostriatal system in mice. Doklady Biol Sci 456(1):160–164. https://doi.org/10.1134/s0012496614030041
Alieva AK, Filatova EV, Kolacheva AA, Rudenok MM, Slominsky PA, Ugrumov MV, Shadrina MI (2016) Transcriptome profile changes in mice with MPTP-induced early stages of Parkinson’s disease. Mol Neurobiol 54(9):6775–6784. https://doi.org/10.1007/s12035-016-0190-y
National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals (2011) The guide for the care and use of laboratory animals, 8th edn. National Academies Press, Washington (DC)
Team RC (2015) R: a language and environment for statistical computing [Internet]. Vienna: R Foundation for Statistical Computing 2014. http://www.R-project.org/. Accessed 09 Aug 2017
Wickham H, Francois R (2015) dplyr: a grammar of data manipulation. http://CRAN.R-project.org/package=dplyr. Accessed 09 Aug 1 2017
Dowle M, Short T, Lianoglou S, Saporta R, Srinivasan A, Antonyan E (2014) data. table: extension of data. frame. http://CRAN.R-project.org/package=data.table. Accessed 09 Aug 2017
Chen H (2016) VennDiagram: generate high-resolution Venn and Euler plots. R package version 1.6. 17. http://CRAN.R-project.org/package=VennDiagram. Accessed 09 Aug 2017
Rasmussen M, Karypis G (2004) gCLUTO: an interactive clustering, visualization, and analysis system. CSE/UMN Technical Rep 04(021):10
Karypis G (2006) gCluto Graphical Clustering Toolkit http://glaros.dtc.umn.edu/gkhome/cluto/gcluto/overview. Accessed 09 Aug 2017
Sturn A, Quackenbush J, Trajanoski Z (2002) Genesis: cluster analysis of microarray data. Bioinformatics (Oxford, England) 18(1):207–208. https://doi.org/10.1093/bioinformatics/18.1.207
Genesis (2002) http://genome.tugraz.at/genesisclient/genesisclient_description.shtml. Accessed 09 Aug 2017
Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4(1):44–57. https://doi.org/10.1038/nprot.2008.211
Huang da W, Sherman BT, Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37(1):1–13. https://doi.org/10.1093/nar/gkn923
DAVID Bioinformatics Resources database (2003) https://david.ncifcrf.gov/summary.jsp. Accessed 09 Aug 2017
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A et al (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43(Database issue):D447–D452. https://doi.org/10.1093/nar/gku1003
STRING database (2003) http://string-db.org. Accessed 17 Mar 2017
Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD (2017) PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 45(D1):D183–d189. https://doi.org/10.1093/nar/gkw1138
PANTHER Classification system http://pantherdb.org. Accessed 17 Mar 2017
Hettne KM, Thompson M, van Haagen HH, van der Horst E, Kaliyaperumal R, Mina E, Tatum Z, Laros JF et al (2016) The implicitome: a resource for rationalizing gene-disease associations. PLoS One 11(2):e0149621. https://doi.org/10.1371/journal.pone.0149621
Pathway Studio https://mammalcedfx.pathwaystudio.com. Accessed 09 Aug 2017
Benjamini Y, Heller R (2008) Screening for partial conjunction hypotheses. Biometrics 64(4):1215–1222. https://doi.org/10.1111/j.1541-0420.2007.00984.x
Gene Ontology database (the framework for the model of biology) (1999) http://www.geneontology.org/. Accessed 09 Aug 2017
Gerlach M, Riederer P (1996) Animal models of Parkinson’s disease: an empirical comparison with the phenomenology of the disease in man. J Neural Transm 103(8–9):987–1041. https://doi.org/10.1007/BF01291788
Przedborski S, Jackson-Lewis V, Djaldetti R, Liberatore G, Vila M, Vukosavic S, Almer G (2000) The parkinsonian toxin MPTP: action and mechanism. Restor Neurol Neurosci 16(2):135–142
Antony PM, Diederich NJ, Kruger R, Balling R (2013) The hallmarks of Parkinson’s disease. FEBS J 280(23):5981–5993. https://doi.org/10.1111/febs.12335
Gandhi S, Muqit MM, Stanyer L et al (2006) PINK1 protein in normal human brain and Parkinson’s disease. Brain 129(7):1720–1731. https://doi.org/10.1093/brain/awl114
Youle RJ, Narendra DP (2011) Mechanisms of mitophagy. Nat Rev Mol Cell Biol 12(1):9–14. https://doi.org/10.1038/nrm3028
Ding WX, Yin XM (2012) Mitophagy: mechanisms, pathophysiological roles, and analysis. Biol Chem 393(7):547–564. https://doi.org/10.1515/hsz-2012-0119
Braak H, Del Tredici K (2004) Poor and protracted myelination as a contributory factor to neurodegenerative disorders. Neurobiol Aging 25(1):19–23. https://doi.org/10.1016/j.neurobiolaging.2003.04.001
Braak H, Del Tredici K (2009) Neuroanatomy and pathology of sporadic Parkinson’s disease. Adv Anat Embryol Cell Biol 201:1–119
Redeker V, Pemberton S, Bienvenut W, Bousset L, Melki R (2012) Identification of protein interfaces between alpha-synuclein, the principal component of Lewy bodies in Parkinson disease, and the molecular chaperones human Hsc70 and the yeast Ssa1p. J Biol Chem 287:32630–32639. https://doi.org/10.1074/jbc.M112.387530
Papagiannakis N, Xilouri M, Koros C, Stamelou M, Antonelou R, Maniati M, Papadimitriou D, Moraitou M et al (2015) Lysosomal alterations in peripheral blood mononuclear cells of Parkinson’s disease patients. Mov Disord 30(13):1830–1834. https://doi.org/10.1002/mds.26433
Nickell JR, Culver JP, Janganati V, Zheng G, Dwoskin LP, Crooks PA (2016) Synthesis and in vitro evaluation of water-soluble 1,4-diphenethylpiperazine analogs as novel inhibitors of the vesicular monoamine transporter-2. Bioorg Med Chem Lett 26(18):4441–4445. https://doi.org/10.1016/j.bmcl.2016.08.001
Volz TJ, Hanson GR, Fleckenstein AE (2006) Measurement of kinetically resolved vesicular dopamine uptake and efflux using rotating disk electrode voltammetry. J Neurosci Methods 155(1):109–115. https://doi.org/10.1016/j.jneumeth.2006.01.002
Lohr KM, Bernstein AI, Stout KA, Dunn AR, Lazo CR, Alter SP, Wang M, Li Y et al (2014) Increased vesicular monoamine transporter enhances dopamine release and opposes Parkinson disease-related neurodegeneration in vivo. Proc Natl Acad Sci U S A 111(27):9977–9982. https://doi.org/10.1073/pnas.1402134111
Wang ZQ, Grigoriadis AE, Mohle-Steinlein U, Wagner EF (1991) A novel target cell for c-fos-induced oncogenesis: development of chondrogenic tumours in embryonic stem cell chimeras. EMBO J 10(9):2437–2450
Wang JD, Cao YL, Li Q, Yang YP, Jin M, Chen D, Wang F, Wang GH et al (2015) A pivotal role of FOS-mediated BECN1/Beclin 1 upregulation in dopamine D2 and D3 receptor agonist-induced autophagy activation. Autophagy 11(11):2057–2073. https://doi.org/10.1080/15548627.2015.1100930
Funding
This work was supported by the Russian Science Foundation (grants no. 16-15-00238 and 17-75-10119).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Experiments with laboratory animals were performed in accordance with The Guide for the Care and Use of Laboratory Animals. The study was approved by the Ethics Committee of the Institute of Molecular Genetics of Russian Academy of Sciences.
Competing Interests
The authors declare that there is no conflict of interest.
Rights and permissions
About this article
Cite this article
Alieva, A., Zyrin, V., Rudenok, M. et al. Whole-Transcriptome Analysis of Mouse Models with MPTP-Induced Early Stages of Parkinson’s Disease Reveals Stage-Specific Response of Transcriptome and a Possible Role of Myelin-Linked Genes in Neurodegeneration. Mol Neurobiol 55, 7229–7241 (2018). https://doi.org/10.1007/s12035-018-0907-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-018-0907-1