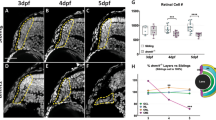
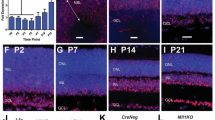
Abstract
Epigenetic modifiers can work in concert with transcription factors to control the transition of cells from proliferating progenitors into quiescent terminally differentiated cells. This transition involves changes in histone methylation and one of the key regulators of this is the H3K4me2/1 histone demethylase LSD1. Here, we show that the highest expression of LSD1 occurs in postmitotic retinal cells during the peak period of rod photoreceptor differentiation. Pharmacological inhibition of LSD1 in retinal explants cultured from PN1 to PN8 had three major effects. It prevented the normal decrease in expression of genes associated with progenitor function, it blocked rod photoreceptor development, and it increased expression of genes associated with other retinal cell types. The maintained expression of progenitor genes was associated with a maintained level of H3K4me2 over the gene and its promoter. Among the genes whose expression was maintained was Hes1, a repressor known to block rod photoreceptor development. The inhibition of rod photoreceptor gene expression occurred in spite of the normal expression of transcription factors CRX and NRL, and the normal accumulation of H3K4me2 marks over the promoter and gene body. We suggest that LSD1 acts in concert with a series of nuclear receptors to modify chromatin structure and repress progenitor genes as well as to inhibit ectopic patterns of gene expression in the differentiating postmitotic retinal cells.










Similar content being viewed by others
References
Kooistra SM, Helin K (2012) Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol 13:297–311
Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941–953
Hou H, Yu H (2010) Structural insights into histone lysine demethylation. Curr Opin Struct Biol 20:739–748
Adamo A, Sese B, Boue S, Castano J, Paramonov I, Barrero MJ, Izpisua Belmonte JC (2011) LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat Cell Biol 13:652–659
McDonald OG, Wu H, Timp W, Doi A, Feinberg AP (2011) Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nat Struct Mol Biol 18:867–874
Kerenyi MA, Shao Z, Hsu YJ, Guo G, Luc S, O’Brien K, Fujiwara Y, Peng C et al (2013) Histone demethylase Lsd1 represses hematopoietic stem and progenitor cell signatures during blood cell maturation. eLife 2, e00633
Sprussel A, Schulte JH, Weber S, Necke M, Handschke K, Thor T, Pajtler KW, Schramm A et al (2012) Lysine-specific demethylase 1 restricts hematopoietic progenitor proliferation and is essential for terminal differentiation. Leukemia 26:2039–2051
Grimaldi P, Pucci M, Di Siena S, Di Giacomo D, Pirazzi V, Geremia R, Maccarrone M (2012) The faah gene is the first direct target of estrogen in the testis: role of histone demethylase LSD1. Cell Mol Life Sci: CMLS 69:4177–4190
Jeon CJ, Strettoi E, Masland RH (1998) The major cell populations of the mouse retina. J Neurosci: Off J Soc Neurosci 18:8936–8946
Zhang SS, Xu X, Liu MG, Zhao H, Soares MB, Barnstable CJ, Fu XY (2006) A biphasic pattern of gene expression during mouse retina development. BMC Dev Biol 6:48
Popova EY, Xu X, DeWan AT, Salzberg AC, Berg A, Hoh J, Zhang SS, Barnstable CJ (2012) Stage and gene specific signatures defined by histones H3K4me2 and H3K27me3 accompany mammalian retina maturation in vivo. PLoS One 7, e46867
Lee MG, Wynder C, Schmidt DM, McCafferty DG, Shiekhattar R (2006) Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem Biol 13(6):563–567
Schmidt DM, McCafferty DG (2007) trans-2-Phenylcyclopropylamine is a mechanism-based inactivator of the histone demethylase LSD1. Biochemistry 46:4408–4416
Shi L, Cui S, Engel JD, Tanabe O (2013) Lysine-specific demethylase 1 is a therapeutic target for fetal hemoglobin induction. Nat Med 19:291–294
Sun G, Alzayady K, Stewart R, Ye P, Yang S, Li W, Shi Y (2010) Histone demethylase LSD1 regulates neural stem cell proliferation. Mol Cell Biol 30:1997–2005
Zhang F, Xu D, Yuan L, Sun Y, Xu Z (2014) Epigenetic regulation of Atrophin1 by lysine-specific demethylase 1 is required for cortical progenitor maintenance. Nat Commun 5:5815
Barnstable CJ (1980) Monoclonal antibodies which recognize different cell types in the rat retina. Nature 286:231–235
Hargrave PA, Adamus G, Arendt A, McDowell JH, Wang J, Szaby A, Curtis D, Jackson RW (1986) Rhodopsin’s amino terminus is a principal antigenic site. Exp Eye Res 42:363–373
Abo T, Balch CM (1981) A differentiation antigen of human NK and K cells identified by a monoclonal antibody (HNK-1). J Immunol 127(3):1024–1029
Naegele JR, Barnstable CJ (1991) A carbohydrate epitope defined by monoclonal antibody VC1.1 is found on N-CAM and other cell adhesion molecules. Brain Res 559(1):118–129
Egelhofer TA, Minoda A, Klugman S, Lee K, Kolasinska-Zwierz P, Alekseyenko AA, Cheung MS, Day DS et al (2011) An assessment of histone-modification antibody quality. Nat Struct Mol Biol 18:91–93
Lobanova ES, Herrmann R, Finkelstein S, Reidel B, Skiba NP, Deng WT, Jo R, Weiss ER et al (2010) Mechanistic basis for the failure of cone transducin to translocate: why cones are never blinded by light. J Neurosci 30(20):6815–6824
Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA (2012) Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 482:221–225
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Popova EY, Grigoryev SA, Fan Y, Skoultchi AI, Zhang SS, Barnstable CJ (2013) Developmentally regulated linker histone H1c promotes heterochromatin condensation and mediates structural integrity of rod photoreceptors in mouse retina. J Biol Chem 288(24):17895–17907
Pinzon-Guzman C, Zhang SS, Barnstable CJ (2011) Specific protein kinase C isoforms are required for rod photoreceptor differentiation. J neurosci 31:18606–18617
Sparrow JR, Hicks D, Barnstable CJ (1990) Cell commitment and differentiation in explants of embryonic rat neural retina. Comparison with the developmental potential of dissociated retina. Brain research. Dev Brain Res 51:69–84
Zhang SS, Fu XY, Barnstable CJ (2002) Tissue culture studies of retinal development. Methods 28:439–447
Zhang SS, Wei J, Qin H, Zhang L, Xie B, Hui P, Deisseroth A, Barnstable CJ et al (2004) STAT3-mediated signaling in the determination of rod photoreceptor cell fate in mouse retina. Invest Ophthalmol Vis Sci 45:2407–2412
Zhang SS, Wei JY, Li C, Barnstable CJ, Fu XY (2003) Expression and activation of STAT proteins during mouse retina development. Exp Eye Res 76:421–431
Siegert S, Cabuy E, Scherf BG, Kohler H, Panda S, Le YZ, Fehling HJ, Gaidatzis D et al (2012) Transcriptional code and disease map for adult retinal cell types. Nat Neurosci 15(3):487–495
Liu MG, Li H, Xu X, Barnstable CJ, Zhang SS (2008) Comparison of gene expression during in vivo and in vitro postnatal retina development. J Ocul Biol Dis Informatics 1:59–72
Tomita K, Ishibashi M, Nakahara K, Ang SL, Nakanishi S, Guillemot F, Kageyama R (1996) Mammalian hairy and enhancer of split homolog 1 regulates differentiation of retinal neurons and is essential for eye morphogenesis. Neuron 16:723–734
Forrest D, Swaroop A (2012) Minireview: the role of nuclear receptors in photoreceptor differentiation and disease. Mol Endocrinol 26:905–915
Yin F, Lan R, Zhang X, Zhu L, Chen F, Xu Z, Liu Y, Ye T et al (2014) LSD1 regulates pluripotency of embryonic stem/carcinoma cells through histone deacetylase 1-mediated deacetylation of histone H4 at lysine 16. Mol Cell Biol 34:158–179
Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, Opravil S, Shiekhattar R et al (2007) p53 is regulated by the lysine demethylase LSD1. Nature 449(7158):105–108
Nicholson TB, Chen T (2009) LSD1 demethylates histone and non-histone proteins. Epigenetics 4(3):129–132
Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, Sun W et al (2009) The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet 41(1):125–129
Choi J, Jang H, Kim H, Kim ST, Cho EJ, Youn HD (2010) Histone demethylase LSD1 is required to induce skeletal muscle differentiation by regulating myogenic factors. Biochem Biophys Res Commun 401:327–332
Musri MM, Carmona MC, Hanzu FA, Kaliman P, Gomis R, Parrizas M (2010) Histone demethylase LSD1 regulates adipogenesis. J Biol Chem 285:30034–30041
Benner C, Konovalov S, Mackintosh C, Hutt KR, Stunnenberg R, Garcia-Bassets I (2013) Decoding a signature-based model of transcription cofactor recruitment dictated by cardinal cis-regulatory elements in proximal promoter regions. PLoS Genet 9, e1003906
Yang M, Gocke CB, Luo X, Borek D, Tomchick DR, Machius M, Otwinowski Z, Yu H (2006) Structural basis for CoREST-dependent demethylation of nucleosomes by the human LSD1 histone demethylase. Mol Cell 23:377–387
Gui H, Li ML, Tsai CC (2011) A tale of tailless. Dev Neurosci 33:1–13
Cui S, Kolodziej KE, Obara N, Amaral-Psarris A, Demmers J, Shi L, Engel JD, Grosveld F et al (2011) Nuclear receptors TR2 and TR4 recruit multiple epigenetic transcriptional corepressors that associate specifically with the embryonic beta-type globin promoters in differentiated adult erythroid cells. Mol Cell Biol 31:3298–3311
Yokoyama A, Takezawa S, Schule R, Kitagawa H, Kato S (2008) Transrepressive function of TLX requires the histone demethylase LSD1. Mol Cell Biol 28:3995–4003
Mulligan P, Yang F, Di Stefano L, Ji JY, Ouyang J, Nishikawa JL, Toiber D, Kulkarni M et al (2011) A SIRT1-LSD1 corepressor complex regulates Notch target gene expression and development. Mol Cell 42:689–699
Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y (2005) Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell 19:857–864
Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA et al (2007) Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature 446:882–887
Garcia-Bassets I, Kwon YS, Telese F, Prefontaine GG, Hutt KR, Cheng CS, Ju BG, Ohgi KA et al (2007) Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell 128:505–518
Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R et al (2005) LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437:436–439
Miyawaki T, Uemura A, Dezawa M, Yu RT, Ide C, Nishikawa S, Honda Y, Tanabe Y et al (2004) Tlx, an orphan nuclear receptor, regulates cell numbers and astrocyte development in the developing retina. J Neurosci 24:8124–8134
Yu RT, Chiang MY, Tanabe T, Kobayashi M, Yasuda K, Evans RM, Umesono K, (2000) The orphan nuclear receptor Tlx regulates Pax2 and is essential for vision. Proceedings of the National Academy of Sciences of the UniteEpigenetic regulation of Atrophin1 by lysine-specific demethylase 1 is required for cortical progenitor maintenance.d States of America 97, 2621–2625
Corbo JC, Cepko CL (2005) A hybrid photoreceptor expressing both rod and cone genes in a mouse model of enhanced S-cone syndrome. PLoS Genet 1, e11
Haider NB, Mollema N, Gaule M, Yuan Y, Sachs AJ, Nystuen AM, Naggert JK, Nishina PM (2009) Nr2e3-directed transcriptional regulation of genes involved in photoreceptor development and cell-type specific phototransduction. Exp Eye Res 89:365–372
Jadhav AP, Mason HA, Cepko CL (2006) Notch 1 inhibits photoreceptor production in the developing mammalian retina. Development 133(5):913–923
Mizeracka K, DeMaso CR, Cepko CL (2013) Notch1 is required in newly postmitotic cells to inhibit the rod photoreceptor fate. Development 140(15):3188–3197
Cheng H, Khan NW, Roger JE, Swaroop A (2011) Excess cones in the retinal degeneration rd7 mouse, caused by the loss of function of orphan nuclear receptor Nr2e3, originate from early-born photoreceptor precursors. Hum Mol Genet 20:4102–4115
Peng GH, Ahmad O, Ahmad F, Liu J, Chen S (2005) The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes. Hum Mol Genet 14:747–764
Hojo M, Ohtsuka T, Hashimoto N, Gradwohl G, Guillemot F, Kageyama R (2000) Glial cell fate specification modulated by the bHLH gene Hes5 in mouse retina. Development 127:2515–2522
Acknowledgments
This research was supported by NEI/NIH grant EY013865 and Macula Vision Research Foundation. We thank Rob Brucklacher from Penn State Hershey Genome Sciences Facility for help with gene expression microarray.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Highlight
1. LSD1 is expressed at transition from late progenitors to rod photoreceptors
2. Under LSD1 inhibition level of H3K4me2 maintained over the progenitor genes
3. LSD1 inhibition prevented the normal decrease in expression of progenitor genes
4. LSD1 inhibition blocked rod photoreceptor development
5. LSD1 inhibition increased other retinal cell type’s genes expression
Evgenya Y. Popova and Carolina Pinzon-Guzman contributed equally to this work.
Rights and permissions
About this article
Cite this article
Popova, E.Y., Pinzon-Guzman, C., Salzberg, A.C. et al. LSD1-Mediated Demethylation of H3K4me2 Is Required for the Transition from Late Progenitor to Differentiated Mouse Rod Photoreceptor. Mol Neurobiol 53, 4563–4581 (2016). https://doi.org/10.1007/s12035-015-9395-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-015-9395-8