Abstract
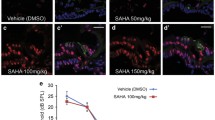
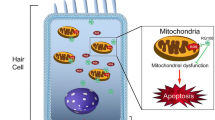
Aminoglycoside-induced hair cell loss is a major cause of hearing impairment in children and deserves more attention in medical research. Epigenetic mechanisms have been shown to protect hair cells from ototoxic drugs. In this study, we focused on the role of dimethylated histone H3K4 (H3K4me2) in hair cell survival. To investigate the effects of lysine-specific demethylase 1 (LSD1)—the histone demethylase primarily responsible for demethylating H3K4me2—on neomycin-induced hair cell loss, isolated cochleae were pretreated with LSD1 inhibitors followed by neomycin exposure. There was a severe loss of hair cells in the organ of Corti after neomycin exposure, and inhibition of LSD1 significantly protected against neomycin-induced hair cell loss. H3K4me2 expression in the nuclei of hair cells decreased after exposure to neomycin, and blocking the decreased expression of H3K4me2 with LSD1 inhibitors prevented hair cell loss. Local delivery of these inhibitors in vivo also protected hair cells from neomycin-induced ototoxicity and maintained the hearing threshold in mice as determined by auditory brain stem response. This inhibition of neomycin-induced apoptosis occurs via reduced caspase-3 activation. Together, our findings demonstrate the protective role for H3K4me2 against neomycin-induced hair cell loss and hearing loss.




Similar content being viewed by others
References
Mills JH, Going JA (1982) Review of environmental factors affecting hearing. Environ Health Perspect 44:119–127
Henley CM, Rybak LP (1995) Ototoxicity in developing mammals. Brain Res Brain Res Rev 20(1):68–90
Schatz A, Bugie E, Waksman SA (2005) Streptomycin, a substance exhibiting antibiotic activity against gram-positive and gram-negative bacteria. Clin Orthop Relat Res 1944(437):3–6
Forge A, Schacht J (2000) Aminoglycoside antibiotics. Audiol Neurootol 5(1):3–22
Perletti G, Vral A, Patrosso MC, Marras E, Ceriani I, Willems P, Fasano M, Magri V (2008) Prevention and modulation of aminoglycoside ototoxicity (review). Mol Med Rep 1(1):3–13
Kooistra SM, Helin K (2012) Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol 13(5):297–311. doi:10.1038/nrm3327
Zhang Y, Reinberg D (2001) Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev 15(18):2343–2360. doi:10.1101/gad.927301
Pattaroni C, Jacob C (2013) Histone methylation in the nervous system: functions and dysfunctions. Mol Neurobiol 47(2):740–756. doi:10.1007/s12035-012-8376-4
Greer EL, Shi Y (2012) Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet 13(5):343–357. doi:10.1038/nrg3173
Rugg-Gunn PJ, Cox BJ, Ralston A, Rossant J (2010) Distinct histone modifications in stem cell lines and tissue lineages from the early mouse embryo. Proc Natl Acad Sci U S A 107(24):10783–10790. doi:10.1073/pnas.0914507107
Yu H, Lin Q, Wang Y, He Y, Fu S, Jiang H, Yu Y, Sun S, Chen Y, Shou J, Li H (2013) Inhibition of H3K9 methyltransferases G9a/GLP prevents ototoxicity and ongoing hair cell death. Cell Death Dis 4:e506. doi:10.1038/cddis.2013.28
Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R (2005) LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437(7057):436–439
Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119(7):941–953. doi:10.1016/j.cell.2004.12.012
Mimasu S, Umezawa N, Sato S, Higuchi T, Umehara T, Yokoyama S (2010) Structurally designed trans-2-phenylcyclopropylamine derivatives potently inhibit histone demethylase LSD1/KDM1. Biochemistry 49(30):6494–6503. doi:10.1021/bi100299r
Gooden DM, Schmidt DM, Pollock JA, Kabadi AM, McCafferty DG (2008) Facile synthesis of substituted trans-2-arylcyclopropylamine inhibitors of the human histone demethylase LSD1 and monoamine oxidases A and B. Bioorg Med Chem Lett 18(10):3047–3051
Binda C, Valente S, Romanenghi M, Pilotto S, Cirilli R, Karytinos A, Ciossani G, Botrugno OA, Forneris F, Tardugno M, Edmondson DE, Minucci S, Mattevi A, Mai A (2010) Biochemical, structural, and biological evaluation of tranylcypromine derivatives as inhibitors of histone demethylases LSD1 and LSD2. J Am Chem Soc 132(19):6827–6833. doi:10.1021/ja101557k
Pekowska A, Benoukraf T, Ferrier P, Spicuglia S (2010) A unique H3K4me2 profile marks tissue-specific gene regulation. Genome Res 20(11):1493–1502. doi:10.1101/gr.109389.110
Zhang J, Parvin J, Huang K (2012) Redistribution of H3K4me2 on neural tissue specific genes during mouse brain development. BMC Genomics 13(Suppl 8):S5. doi:10.1186/1471-2164-13-S8-S5
Popova EY, Xu X, DeWan AT, Salzberg AC, Berg A, Hoh J, Zhang SS, Barnstable CJ (2012) Stage and gene specific signatures defined by histones H3K4me2 and H3K27me3 accompany mammalian retina maturation in vivo. PLoS One 7(10):e46867. doi:10.1371/journal.pone.0046867
Forge A, Richardson G (1993) Freeze fracture analysis of apical membranes in cochlear cultures: differences between basal and apical-coil outer hair cells and effects of neomycin. J Neurocytol 22(10):854–867
Richardson GP, Russell IJ (1991) Cochlear cultures as a model system for studying aminoglycoside induced ototoxicity. Hear Res 53(2):293–311
Gale JE, Marcotti W, Kennedy HJ, Kros CJ, Richardson GP (2001) FM1-43 dye behaves as a permeant blocker of the hair-cell mechanotransducer channel. J Neurosci 21(18):7013–7025
Jia S, Yang S, Guo W, He DZ (2009) Fate of mammalian cochlear hair cells and stereocilia after loss of the stereocilia. J Neurosci 29(48):15277–15285
Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ (2004) Epigenetic programming by maternal behavior. Nat Neurosci 7(8):847–854. doi:10.1038/nn1276
Suva ML, Riggi N, Bernstein BE (2013) Epigenetic reprogramming in cancer. Science 339(6127):1567–1570. doi:10.1126/science.1230184
Roidl D, Hacker C (2014) Histone methylation during neural development. Cell Tissue Res 356(3):539–552. doi:10.1007/s00441-014-1842-8
Papait R, Cattaneo P, Kunderfranco P, Greco C, Carullo P, Guffanti A, Vigano V, Stirparo GG, Latronico MV, Hasenfuss G, Chen J, Condorelli G (2013) Genome-wide analysis of histone marks identifying an epigenetic signature of promoters and enhancers underlying cardiac hypertrophy. Proc Natl Acad Sci U S A 110(50):20164–20169. doi:10.1073/pnas.1315155110
Falk M, Lukasova E, Stefancikova L, Baranova E, Falkova I, Jezkova L, Davidkova M, Bacikova A, Vachelova J, Michaelidesova A, Kozubek S (2014) Heterochromatinization associated with cell differentiation as a model to study DNA double strand break induction and repair in the context of higher-order chromatin structure. Applied Radiat Isot 83 Pt B:177–185. doi:10.1016/j.apradiso.2013.01.029
Gospodinov A, Herceg Z (2013) Chromatin structure in double strand break repair. DNA repair 12(10):800–810. doi:10.1016/j.dnarep.2013.07.006
Maze I, Noh KM, Allis CD (2013) Histone regulation in the CNS: basic principles of epigenetic plasticity. Neuropsychopharmacoly 38(1):3–22. doi:10.1038/npp.2012.124
Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, Paylor RE, Lubin FD (2010) Histone methylation regulates memory formation. J Neurosci 30(10):3589–3599. doi:10.1523/JNEUROSCI.3732-09.2010
Kuzumaki N, Ikegami D, Tamura R, Hareyama N, Imai S, Narita M, Torigoe K, Niikura K, Takeshima H, Ando T, Igarashi K, Kanno J, Ushijima T, Suzuki T (2011) Hippocampal epigenetic modification at the brain-derived neurotrophic factor gene induced by an enriched environment. Hippocampus 21(2):127–132. doi:10.1002/hipo.20775
Huang HS, Matevossian A, Whittle C, Kim SY, Schumacher A, Baker SP, Akbarian S (2007) Prefrontal dysfunction in schizophrenia involves mixed-lineage leukemia 1-regulated histone methylation at GABAergic gene promoters. J Neurosci 27(42):11254–11262
Orford K, Kharchenko P, Lai W, Dao MC, Worhunsky DJ, Ferro A, Janzen V, Park PJ, Scadden DT (2008) Differential H3K4 methylation identifies developmentally poised hematopoietic genes. Dev Cell 14(5):798–809. doi:10.1016/j.devcel.2008.04.002
Kim T, Buratowski S (2009) Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell 137(2):259–272. doi:10.1016/j.cell.2009.02.045
Faucher D, Wellinger RJ (2010) Methylated H3K4, a transcription-associated histone modification, is involved in the DNA damage response pathway. PLoS Genet 6(8). doi:10.1371/journal.pgen.1001082
Tyagi S, Herr W (2009) E2F1 mediates DNA damage and apoptosis through HCF-1 and the MLL family of histone methyltransferases. EMBO J 28(20):3185–3195. doi:10.1038/emboj.2009.258
Bergmann JH, Rodriguez MG, Martins NM, Kimura H, Kelly DA, Masumoto H, Larionov V, Jansen LE, Earnshaw WC (2011) Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. The EMBO journal 30(2):328–340. doi:10.1038/emboj.2010.329
Nightingale KP, Gendreizig S, White DA, Bradbury C, Hollfelder F, Turner BM (2007) Cross-talk between histone modifications in response to histone deacetylase inhibitors: MLL4 links histone H3 acetylation and histone H3K4 methylation. J Biol Chem 282(7):4408–4416. doi:10.1074/jbc.M606773200
Jiang H, Sha SH, Forge A, Schacht J (2006) Caspase-independent pathways of hair cell death induced by kanamycin in vivo. Cell Death Differ 13(1):20–30
Matsui JI, Haque A, Huss D, Messana EP, Alosi JA, Roberson DW, Cotanche DA, Dickman JD, Warchol ME (2003) Caspase inhibitors promote vestibular hair cell survival and function after aminoglycoside treatment in vivo. J Neurosci 23(14):6111–6122
Cunningham LL, Cheng AG, Rubel EW (2002) Caspase activation in hair cells of the mouse utricle exposed to neomycin. J Neurosci 22(19):8532–8540
Taylor RR, Nevill G, Forge A (2008) Rapid hair cell loss: a mouse model for cochlear lesions. J Assoc Res Otolaryngol 9(1):44–64. doi:10.1007/s10162-007-0105-8
Selimoglu E (2007) Aminoglycoside-induced ototoxicity. Curr Pharm Des 13(1):119–126
Acknowledgments
The authors would like to thank Min Yu and Shaoyang Sun for their technical assistance and Yalin Huang for help with the confocal microscope. This work was supported by grants from the Major State Basic Research Development Program of China (973 Program) (2011CB504506, 2010CB945503) and the National Natural Science Foundation of China (Nos. 81070793, 81230019, 81300825), the Program for Changjiang Scholars and Innovative Research Team in University (IRT1010), the Specialized Research Fund for the Doctor Program of Higher Education (20120071110077), the Program of Outstanding Shanghai Academic Leader (11XD1401300), the Program of Leading Medical Personnel in Shanghai, the Fundamental Research Funds for the Central Universities (2242014R30022, NO2013WSN085), and the China Postdoctoral Science Foundation Funded Project (No. 2014M551328)
Conflict of Interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Yingzi He, Huiqian Yu, and Chengfu Cai contributed equally to this work.
Rights and permissions
About this article
Cite this article
He, Y., Yu, H., Cai, C. et al. Inhibition of H3K4me2 Demethylation Protects Auditory Hair Cells from Neomycin-Induced Apoptosis. Mol Neurobiol 52, 196–205 (2015). https://doi.org/10.1007/s12035-014-8841-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-014-8841-3