Abstract
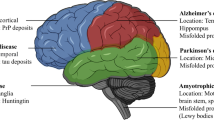
Neurodegenerative foldopathies are characterized by aberrant folding of diseased modified proteins, which are major constituents of the intracellular and extracellular lesions. These lesions correlate with the cognitive and/or motor impairment seen in these diseases. The majority of the disease modified proteins in neurodegenerative foldopathies belongs to the group of proteins termed as intrinsically disordered proteins (IDPs). Several independent studies have showed that abnormal protein processing constitutes the key pathological feature of these disorders. The current review focuses on protein truncation as a common denominator of neurodegenerative foldopathies, which is considered to be the major driving force behind the pathological metamorphosis of brain IDPs. The aim of the review is to emphasize the key role of the protein truncation in the pathogenic pathways of neurodegenerative diseases. A deeper understanding of the complex downstream processing of the IDPs, resulting in the generation of pathologically modified proteins might be a prerequisite for the successful therapeutic strategies of several fatal neurodegenerative diseases.


Similar content being viewed by others
References
Skrabana R, Skrabanova-Khuebachova M, Kontsek P, Novak M (2006) Alzheimer’s-disease-associated conformation of intrinsically disordered tau protein studied by intrinsically disordered protein liquid-phase competitive enzyme-linked immunosorbent assay. Analytical biochem 359:230–237
Sticht H, Bayer P, Willbold D, Dames S, Hilbich C et al (1995) Structure of amyloid A4-(1–40)-peptide of Alzheimer’s disease. Eur J Biochem 233:293–298
Dyson HJ, Wright PE (2005) Intrisically unstructured proteins and their functions. Nat Rev Mol Cell Biol 6:197–208
Weinreb PH, Zhen W, Poon AW, Conway KA, Lansbury PT Jr (1996) NACP, a protein implicated in Alzheimer’s disease and learning, is natively unfolded. Biochemistry 35:13709–13715
Raychaudhuri S, Majumder P, Sarkar S, Giri K, Mukhopadhyay D, Bhattacharyya NP (2008) Huntingtin interacting protein HYPK is intrinsically unstructured. Proteins 71:1686–1698
Chen YW (2003) Local protein unfolding and pathogenesis of polyglutamine-expansion diseases. PROTEINS: Struct Funct Genet 51:68–73
He B, Wang K, Li Y, Xue B, Uversky VN, Dunker AK (2009) Predicting intrinsic disorder in proteins: An overview. Cell Res 19:929–949
Uversky VN, Oldfield CJ, Midic U, Xie H, Xue B et al (2009) Unfoldomics of human diseases: Linking protein intrinsic disorder with diseases. BMC Genomics 10(Suppl 1):S7. doi:10.1186/1471-2164-10-S1-S7
Wischik CM, Novak M, Edwards PC, Klug A, Tichelaar W, Crowther RA (1988) Structural characterization of the core of the paired helical filament of Alzheimer disease. Proc Natl Acad Sci USA 85:4884–4888
Wischik CM, Novak M, Thøgersen HC, Edwards PC, Runswick MJ et al (1988) Isolation of a fragment of tau derived from the core of the paired helical filament of Alzheimer disease. Proc Natl Acad Sci USA 85:4506–4510
Novak M, Kabat J, Wischik CM (1993) Molecular characterization of the minimal protease resistant tau unit of the Alzheimer’s disease paired helical filament. EMBO J 12:365–370
Novak M (1994) Truncated tau protein as a new marker for Alzheimer’s disease. Acta Virol 38:173–189
Landles C, Sathasivam K, Weiss A, Woodman B, Moffitt H et al (2010) Proteolysis of mutant huntingtin produces an exon 1 fragment that accumulates as an aggregated protein in neuronal nuclei in Huntington disease. J Biol Chem 285:8808–8823
Lunkes A, Lindenberg KS, Ben-Haiem L, Weber C, Devys D, Landwehrmeyer GB, Mandel JL, Trottier Y (2002) Proteases acting on mutant huntingtin generate cleaved products that differentially build up cytoplasmic and nuclear inclusions. Mol Cell 10:259–269
Grundke-Iqbal I, Iqbal K, Tung YC, Quinlan M, Wisniewski HM, Binder LI (1986) Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc Natl Acad Sci U S A 83:4913–4917
Schilling B, Gafni J, Torcassi C, Cong X, Row RH et al (2006) Huntingtin phosphorylation sites mapped by mass spectrometry. Modulation of cleavage and toxicity. J Biol Chem 281:23686–23697
Fei E, Jia N, Zhang T, Ma X, Wang H et al (2007) Phosphorylation of ataxin-3 by glycogen synthase kinase 3beta at serine 256 regulates the aggregation of ataxin-3. Biochem Biophys Res Commun 357:487–492
Arai T, Hasegawa M, Nonoka T, Kametani F, Yamashita M et al (2010) Phosphorylated and cleaved TDP-43 in ALS, FTLD and other neurodegenerative disorders and in cellular models of TDP-43 proteinopathy. Neuropathology 30:170–181
Bondareff W, Wischik CM, Novak M, Amos WB, Klug A, Roth M (1990) Molecular analysis of neurofibrillary degeneration in Alzheimer’s disease. An immunohistochemical study. Am J Pathol 137:711–723
Kalchman MA, Graham RK, Xia G, Koide HB, Hodgson JG et al (1996) Huntingtin is ubiquitinated and interacts with a specific ubiquitin- conjugating enzyme. J Biol Chem 271:19385–19394
Poukka H, Karvonen U, Janne OA, Palvimo JJ (2000) Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1). Proc Natl Acad Sci USA 97:14145–14150
de Pril R, Fischer DF, Roos RA, van Leeuwen FW (2007) Ubiquitin-conjugating enzyme E2–25 K increases aggregate formation and cell death in polyglutamine diseases. Mol Cell Neurosci 34:10–19
Maynard CJ, Böttcher C, Ortega Z, Smith R, Florea BI et al (2009) Accumulation of ubiquitin conjugates in a polyglutamine disease model occurs without global ubiquitin/proteasome system impairment. Proc Natl Acad Sci U S A 106:13986–13991
Sasaki N, Fukatsu R, Tsuzuki K, Hayashi Y, Yoshida T et al (1998) Advanced glycation end products in Alzheimer’s disease and other neurodegenerative diseases. Am J Pathol 153:1149–1155
Ma L, Nicholson LF (2004) Expression of the receptor for advanced glycation end products in Huntington’s disease caudate nucleus. Brain Res 1018:10–17
Padmaraju V, Bhaskar JJ, Prasada Rao UJ, Salimath PV, Rao KS (2011) Role of advanced glycation on aggregation and DNA binding properties of α-synuclein. J Alzheimers Dis 24(Suppl 2):211–221
Wang JZ, Grundke-Iqbal I, Iqbal K (1996) Glycosylation of microtubule-associated protein tau: An abnormal posttranslational modification in Alzheimer’s disease. Nat Med 2:871–875
Marz P, Stetefeld J, Bendfeldt K, Nitsch C, Reinstein J et al (2006) Ataxin-10 interacts with O-linked beta-N-acetylglucosamine transferase in the brain. J Biol Chem 281:20263–20270
Horiguchi T, Uryu K, Giasson BI, Ischiropoulos H, LightFoot R et al (2003) Nitration of tau protein is linked to neurodegeneration in tauopathies. Am J Pathol 163:1021–1031
Yu Z, Xu X, Xiang Z, Zhou J, Zhang Z, Hu C, He C (2010) Nitrated alpha-synuclein induces the loss of dopaminergic neurons in the substantia nigra of rats. PLoS One 5:e9956
Steffan JS, Agrawal N, Pallos J, Rockabrand E, Trotman LC et al (2004) SUMO modification of huntingtin and Huntington’s disease pathology. Science 304:100–104
Riley BE, Zoghbi HY, Orr HT (2005) SUMOylation of the polyglutamine repeat protein, ataxin-1, is dependent on a functional nuclear localization signal. J Biol Chem 280:21942–21948
Dorval V, Fraser PE (2006) Small ubiquitin-like modifier (SUMO) modification of natively unfolded proteins tau and alpha-synuclein. J Biol Chem 281:9919–9924
Kim YM, Jang WH, Quezado MM, Oh Y, Chung KC, Junn E, Mouradian MM (2011) Proteasome inhibition induces α-synuclein SUMOylation and aggregate formation. J Neurol Sci 307:157–161
Harper PS, Youngman S, Anderson MA, Sarfarazi M, Quarrell O, Tanzi R et al (1985) Genetic linkage between Huntington’s disease and the DNA polymorphism G8 in South Wales families. J Med Genet 22:447–450
Yoshioka K, Miki T, Katsuya T, Ogihara T, Sakaki Y (1991) The 717Val–Ile substitution in amyloid precursor protein is associated with familial Alzheimer’s disease regardless of ethnic groups. Biochem Biophys Res Commun 178:1141–1146
Wszolek ZK, Cordes M, Calne DB, Münter MD, Cordes I, Pfeifer RF (1993) Hereditary Parkinson disease: Report of 3 families with dominant autosomal inheritance. Nervenarzt 64:331–335
Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A et al (1997) Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science 276:2045–2047
Spillantini MG, Murrell JR, Goedert M, Farlow MR, Klug A, Ghetti B (1998) Mutation in the tau gene in familial multiple system tauopathy with presenile dementia. Proc Natl Acad Sci U S A 95:7737–7741
Kruger R, Kuhn W, Muller T, Woitalla D, Graeber M, Kosel S, Przuntek H, Epplen JT, Schols L, Riess O (1998) Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson’s disease. Nat Genet 18:106–108
Zareparsi S, Kaye J, Camicioli R, Kramer P, Nutt J, Bird T, Litt M, Payami H (1998) Analysis of the alpha-synuclein G209A mutation in familial Parkinson’s disease. Lancet 351:37–38
De Jonghe C, Esselens C, Singh SK, Craessaerts K, Serneels S et al (2001) Pathogenic APP mutations near the γ-secretase cleavage site differentially affect Aβ secretion and APP C-terminal fragment stability. Hum Mol Genet 10:1665–1671
Squitieri F, Gellera C, Cannella M, Mariotti C, Cislaghi G et al (2003) Homozygosity for CAG mutation in Huntington disease is associated with a more severe clinical course. Brain 126:946–955
Kovacech B, Skrabana R, Novak M (2010) Transition of tau protein from disordered to misordered in Alzheimer’s disease. Neurodegener Dis 7:24–27
Cente M, Filipcik P, Pevalova M, Novak M (2006) Expression of a truncated tau protein induces oxidative stress in a rodent model of tauopathy. Eur J Neurosci 24:1085–1090
Quintanilla RA, Matthews-Roberson TA, Dolan PJ, Johnson GV (2009) Caspase-cleaved tau expression induces mitochondrial dysfunction in immortalized cortical neurons: Implications for the pathogenesis of Alzheimer disease. J Biol Chem 284:18754–18766
Amadoro G, Corsetti V, Stringaro A, Colone M, D’Aguanno S et al (2010) A NH2 tau fragment targets neuronal mitochondria at AD synapses: Possible implications for neurodegeneration. J Alzheimers Dis 21:445–470
Siman R, Card JP, Davis LG (1990) Proteolytic processing β-amyloid precursor by calpain I. J Neurosci 70:2400–2411
Nieto A, Correas I, López-Otín C, Avila J (1991) Tau-related protein present in paired helical filaments has a decreased tubulin binding capacity as compared with microtubule-associated protein tau. Biochim Biophys Acta 1096:197–204
Zilka N, Filipcik P, Koson P, Fialova L, Skrabana R et al (2006) Truncated tau from sporadic Alzheimer’s disease suffices to drive neurofibrillary degeneration in vivo. FEBS Lett 580:3582–3588
Galvan V, Gorostiza OF, Banwait S, Ataie M, Logvinova AV et al (2006) Reversal of Alzheimer’s-like pathology and behavior in human APP transgenic mice by mutation of Asp664. Proc Natl Acal Sci 103:7130–7135
Liu CW, Giasson BI, Lewis KA, Lee VM, Demartino GN, Thomas PJ (2005) A precipitating role for truncated alpha-synuclein and the proteasome in alpha-synuclein aggregation: Implications for pathogenesis of Parkinson’s disease. J Biol Chem 280:22670–22678
Dufty BM, Warner LR, Hou ST, Jiang SX, Gomez-Isla T et al (2007) Calpain-cleavage of α-synuclein connecting proteolytic processing to disease-linked aggregation. Am J Pathol 170:1725–1738
DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N (1997) Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277:1990–1993
Hackam AS, Singaraja R, Wellington CL, Metzler M, McCutcheon K, Zhang T, Kalchman M, Hayden MR (1998) The influence of huntingtin protein size on nuclear localization and cellular toxicity. J Cell Biol 141:1097–1105
Hackam AS, Singaraja R, Zhang T, Gan L, Hayden MR (1999) In vitro evidence for both the nucleus and cytoplasm as subcellular sites of pathogenesis in Huntington’s disease. Hum Mol Genet 8:25–33
Wellington CL, Singaraja R, Ellerby L, Savill J, Roy S et al (2000) Inhibiting caspase cleavage of huntingtin reduces toxicity and aggregate formation in neuronal and nonneuronal cells. J Biol Chem 275:19831–19838
Mende-Mueller LM, Toneff T, Hwang SR, Chesselet MF, Hook VY (2001) Tissue-specific proteolysis of Huntingtin (htt) in human brain: Evidence of enhanced levels of N- and C-terminal htt fragments in Huntington’s disease striatum. J Neurosci 21:1830–1837
Gafni J, Ellerby LM (2002) Calpain activation in Huntington’s disease. J Neurosci 22:4842–4849
Ikeda H, Yamaguchi M, Sugai S, Aze Y, Narumiya S, Kakizuka A (1996) Expanded polyglutamine in the Machado–Joseph disease protein induces cell death in vitro and in vivo. Nat Genet 13:196–202
Goti D, Katzen SM, Mez J, Kurtis N, Kiluk J et al (2004) A mutant ataxin-3 putative-cleavage fragment in brains of Machado–Joseph disease patients and transgenic mice is cytotoxic above a critical concentration. J Neurosci 24:10266–10279
Haacke AF, Hartl U, Breuer P (2007) Calpain inhibition is sufficient to suppress aggregation of polyglutamine-expanded ataxin-3. J Biol Chem 282:18851–18856
Young JE, Gouw L, Propp S, Sopher BL, Taylor J et al (2007) Proteolytic cleavage of ataxin-7 by caspase-7 modulates cellular toxicity and transcriptional dysregulation. J Biol Chem 282:30150–30160
Arai T, Ikeda K, Akiyama H, Nonaka T, Hasegawa M et al (2004) Identification of amino-terminally cleaved tau fragments that distinguish progressive supranuclear palsy from corticobasal degeneration. Ann Neurol 55:72–79
Mondragón-Rodríguez S, Mena R, Binder LI, Smith MA, Perry G, García-Sierra F (2008) Conformational changes and cleavage of tau in Pick bodies parallel the early processing of tau found in Alzheimer pathology. Neuropathol Appl Neurobiol 34:62–75
Nonaka T, Kametani F, Arai T, Akiyama H, Hasegawa M (2009) Truncation and pathogenic mutations facilitate the formation of intracellular aggregates of TDP-43. Hum Mol Genet 18:3353–3364
Guillozet-Bongaarts AL, Garcia-Sierra F, Reynolds MR, Horowitz PM et al (2005) Tau truncation during neurofibrillary tangle evolution in Alzheimer’s disease. Neurobiol Aging 26:1015–1022
Zhou A, Webb G, Zhu X, Steiner DF (1999) Proteolysis processing in the secretory pathway. J Biol Chem 274:20745–20748
Novak M, Wischik CM, Edwards PC, Panell R, Milstein C (1989) Characterization of the first monoclonal antibody against the pronase resistant core of the Alzheimer PHF. Prog Clin Biol Res 317:755–761
Novak M, Jakes R, Edwards PC, Milstein C, Wischik CM (1991) Difference between the tau protein of Alzheimer paired helical filament core and normal tau revealed by epitope analysis of monoclonal antibodies 423 and 7.51. Proc Natl Acad Sci USA 88:5837–5841
Koson P, Zilka N, Kovac A, Kovacech B, Korenova M, Filipcik P, Novak M (2008) Truncated tau expression levels determine life span of a rat model of tauopathy without causing neuronal loss or correlating with terminal neurofibrillary tangle load. Eur J Neurosci 28:239–246
Filipcik P, Zilka N, Bugos O, Kucerak J, Koson P, Novak P, Novak M (2012) First transgenic rat model developing progressive cortical neurofibrillary tangles. Neurobiol Aging 33:1448–1456
Abraha A, Ghoshal N, Gamblin TC, Cryns V, Berry RW, Kuret J, Binder LI (2000) C-terminal inhibition of tau assembly in vitro and in Alzheimer’s disease. J Cell Sci 113(Pt 21):3737–3745
Gamblin TC, Chen F, Zambrano A, Abraha A, Lagalwar S et al (2003) Caspase cleavage of tau: Linking amyloid and neurofibrillary tangles in Alzheimer’s disease. Proc Natl Acad Sci USA 100:10032–10037
Rissman RA, Poon WW, Blurton-Jones M, Oddo S, Torp R et al (2004) Caspase-cleavage of tau is an early event in Alzheimer disease tangle pathology. J Clin Invest 114:121–130
Cotman CW, Poon WW, Rissman RA, Blurton-Jones M (2005) The role of caspase cleavage of tau in Alzheimer disease neuropathology. J Neuropathol Exp Neurol 64:104–112
deCalignon A, Fox LM, Pitstick R, Carlson GA, Bacskai BJ, Spires-Jones TL, Hyman BT (2010) Caspase activation precedes and leads to tangles. Nature 464:1201–1204
Spires-Jones TL, de Calignon A, Matsui T, Zehr C, Pitstick R et al (2008) In vivo imaging reveals dissociation between caspase activation and acute neuronal death in tangle-bearing neurons. J Neurosci 28:862–867
Amadoro G, Serafino AL, Barbato C, Ciotti MT, Sacco A, Calissano P, Canu N (2004) Role of N-terminal tau domain integrity on the survival of cerebellar granule neurons. Cell Death Differ 11:217–230
Zilkova M, Zilka N, Kovac A, Kovacech B, Skrabana R, Skrabanova M, Novak M (2011) Hyperphosphorylated truncated protein tau induces caspase-3 independent apoptosis-like pathway in the Alzheimer’s disease cellular model. J Alzheimers Dis 23:161–169
Hoogeveen AT, Willemsen R, Meyer N, de Rooij KE, Roos RA, van Ommen GJ, Galjaard H (1993) Characterization and localization of the Huntington disease gene product. Hum Mol Genet 2:2069–2073
Becher MW, Kotzuk JA, Sharp AH, Davies SW, Bates GP, Price DL, Ross C (1998) Intranuclear neuronal inclusions in Huntington’s disease and dentatorubral and pallidoluysian atrophy: Correlation between the density of inclusions and IT15 CAG triplet repeat length. Neurobiol Dis 4:387–397
Wellington CL, Ellerby LM, Hackam AS, Margolis RL, Trifiro MA et al (1998) Caspase cleavage of gene products associated with triplet expansion disorders generates truncated fragments containing the polyglutamine tract. J Biol Chem 273:9158–6917
Sieradzan KA, Mechan AO, Jones L, Wanker EE, Nukina N, Mann DM (1999) Huntington’s disease intranuclear inclusions contain truncated, ubiquitinated huntingtin protein. Exp Neurol 156:92–99
Lee JA, Lim CS, Lee SH, Kim H, Nukina N, Kaang BK (2003) Aggregate formation and the impairment of long-term synaptic facilitation by ectopic expression of mutant huntingtin in Aplysia neurons. J Neurochem 85:160–169
Martindale D, Hackam A, Wieczorek A, Ellerby L, Wellington C et al (1998) Length of huntingtin and its polyglutamine tract influences localization and frequency of intracellular aggregates. Nat Genet 18:150–154
Cooper JK, Schilling G, Peters MF, Herring WJ, Sharp AH et al (1998) Truncated N-terminal fragments of huntingtin with expanded glutamine repeats form nuclear and cytoplasmic aggregates in cell culture. Hum Mol Genet 7:783–790
Gervais F, Xu D, Robertson G, Vaillancourt J, Zhu Y et al (1999) Involvement of caspases in proteolytic cleavage of Alzheimer’s b-amyloid precursor protein and amyloidogenic b-peptide formation. Cell 97:395–406
Selkoe DJ (2008) Biochemistry and molecular biology of amyloid beta-protein and the mechanism of Alzheimer’s disease. Handb Clin Neurol 89:245–260
Chow VW, Mattson MP, Wong PC, Gleichmann M (2010) An overview of APP processing enzymes and products. Neuromol Med 12:1–12
Devi L, Prabhu BM, Galati DF, Avadhani NG, Anandatheerthavarada HK (2006). Accumulation of amyloid precursor protein in the mitochondrial import channels of human Alzheimer's disease brain is associated with mitochondrial dysfunction. J Neurosci 26:9057–9068
Baba M, Nakajo S, Tu PH, Tomita T, Nakaya K, Lee VM, Trojanowski JQ, Iwatsubo T (1998) Aggregation of alpha-synuclein in Lewy bodies of sporadic Parkinson’s disease and dementia with Lewy bodies. Am J Pathol 152:879–884
Anderson JP, Walker DE, Goldstein JM, de Laat R, Banducci K et al (2000) Phosphorylation of Ser-129 is the dominant pathological modification of alpha-synuclein in familial and sporadic Lewy body disease. J Biol Chem 281:29739–29752
Campbell BC, McLean CA, Culvenor JG, Gai WP, Blumbergs PC et al (2001) The solubility of alpha-synuclein in multiple system atrophy differs from that of dementia with Lewy bodies and Parkinson’s disease. J Neurochem 76:87–96
Hoyer W, Cherny D, Subramaniam V, Jovin TM (2004) Impact of the acidic C-terminal region comprising amino acids 109–140 on alphasynuclein aggregation in vitro. Biochemistry 43:16233–16242
Sung JY, Park SM, Lee CH, Um JW, Lee HJ et al (2005) Proteolytic cleavage of extracellular secreted α-synuclein via matrix metalloproteinases. J Biol Chem 280:25216–25224
Li W, West N, Colla E, Pletnikova O, Troncoso JC et al (2005) Aggregation promoting C-terminal truncation of alpha-synuclein is a normal cellular process and is enhanced by the familial Parkinson’s disease-linked mutations. Proc Natl Acad Sci USA 102:2162–2167
Levitan K, Chereau D, Cohen SI, Knowles TP, Dobson CM et al (2011) Conserved C-terminal charge exerts a profound influence on the aggregation rate of α-synuclein. J Mol Biol 411:329–333
Michell AW, Tofaris GK, Gossage H, Tyers P, Spillantini MG, Barker RA (2007) The effect of truncated human alpha-synuclein (1–120) on dopaminergic cells in a transgenic mouse model of Parkinson’s disease. Cell Transplant 16:461–474
Daher JPL, Ying M, Banerjee R, McDonald RS, Hahn MD et al (2009) Conditional transgenic mice expressing C-terminally truncated human α-synuclein (αSyn119) exhibit reduced striatal dopamine without loss of nigrostriatal pathway dopaminergic neurons. Mol Neurodegener 4:34. doi:10.1186/1750-1326-4-34
Serpell LC, Berriman J, Jakes R, Goedert M, Crowther RA (2000) Fiber diffraction of synthetic α-synuclein filaments shows amyloid-like cross-β conformation. Proc Natl Acad Sci USA 97:4897–4902
Igaz LM, Kwong LK, Xu Y, Truax AC, Uryu K, Neumann M et al (2008) Enrichment of C-terminal fragments in TAR DNA-binding protein-43 cytoplasmic inclusions in brain but not in spinal cord of frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Am J Pathol 173:182–194
Igaz LM, Kwong LK, Chen-Plotkin A, Winton MJ, Unger TL et al (2009) Expression of TDP-43 C-terminal fragments in vitro recapitulates pathological features of TDP-43 proteinopathies. J Biol Chem 284:8516–8524
Timmer JC, Salvesan GS (2007) Caspase substrates. Cell Death Differ 14:66–72
Tompa P et al (2004) On the sequential determinants of calpain cleavage. J Biol Chem 279:20775–20785
Gafni J, Hermel E, Young JE, Wellington CL, Hayden MR, Ellerby LM (2004) Inhibition of calpain cleavage of huntingtin reduces toxicity: Accumulation of calpain/caspase fragments in the nucleus. J Biol Chem 279:20211–20220
Yang LS, Ksiezak-Reding H (1995) Calpain induced proteolysis of normal human tau and tau associated with paired helical filaments. Eur J Biochem 233:9–17
Kim HJ, Lee D, Lee CH, Chung KC, Kim J, Paik SR (2006) Calpain-resistant fragment(s) of alpha-synuclein regulates the synuclein-cleaving activity of 20S proteasome. Arch Biochem Biophy 455:40–47
Guillozet-Bongaarts AL, Glajch KE, Libson EG, Cahill ME, Bigio E, Berry RW, Binder LI (2007) Phosphorylation and cleavage of tau in non-AD tauopathies. Acta Neuropathol 113:513–520
Schilling G, Wood JD, Duan K, Slunt HH, Gonzales V et al (1999) Nuclear accumulation of truncated atrophin-1 fragments in a transgenic mouse model of DRPLA. Neuron 24:275–286
Sydow A, Van der Jeugd A, Zheng F, Ahmed T, Balschun D, Petrova O, Drexler D, Zhou L, Rune G, Mandelkow E, D’Hooge R, Alzheimer C, Mandelkow EM (2011) Tau-induced defects in synaptic plasticity, learning, and memory are reversible in transgenic mice after switching off the toxic Tau mutant. J Neurosci 31:2511–2525
Gil JM, Rego AC (2008) Mechanisms of neurodegeneration in Huntington’s disease. Eur J Neurosci 27:2803–2820
Warby SC, Doty CN, Graham RK, Carroll JB, Yang YZ, Singaraja RR, Overall CM, Hayden MR (2008) Activated caspase-6 and caspase-6-cleaved fragments of huntingtin specifically colocalize in the nucleus. Hum Mol Genet 17:2390–2404
Saudou F, Finkbeiner S, Devys D, Greenberg ME (1998) Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell 95:55–66
Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S (2004) Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431:805–810
Xu S, Zhou M, Yu S, Cai Y, Zhang A, Uéda K, Chan P (2006) Oxidative stress induces nuclear translocation of C-terminus of alpha-synuclein in dopaminergic cells. Biochem Biophys Res Commun 342:330–335
Neumann M, Sampathu DM, Kwong LK, Truax AD, Micsenyi MC et al (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314:130–133
Pesiridis GS, Tripathy K, Tanik S, Trojanowski JQ, Lee VM (2011) A “two-hit” hypothesis for inclusion formation by carboxyl-terminal fragments of TDP-43 protein linked to RNA depletion and impaired microtubule-dependent transport. J Biol Chem 286:18845–18855
Perez MK, Paulson HL, Pendse SJ, Saionz SJ, Bonini NM, Pittman RN (1998) Recruitment and the role of nuclear localization in polyglutamine-mediated aggregation. J Cell Biol 143:1457–1470
Kenessey A, Nacharaju P, Ko LW, Yen SH (1997) Degradation of tau by lysosomal enzyme cathepsin D: Implication for Alzheimer neurofibrillary degeneration. J Neurochem 69:2026–2038
Zhang L, Sheng R, Qin Z (2009) The lysozome and neurodegenerative diseases. Acta Biochim Biophys Sin 41:437–445
Johnson GV, Jope RS, Binder LI (1989) Proteolysis of tau by calpain. Biochem Biophys Res Commun 29:1505–1511
Huang Y, Wang KK (2001) The calpain family and human disease. Trends Mol Med 7:355–362
Wang KK, Villalobo A, Roufogalis BD (1989) Calmodulin-binding proteins as calpain substrates. J Biol Chem 262:693–706
Pariat M et al (2001) The sensitivity of c-Jun and c-Fos proteins to calpains depends on conformational determinants of the monomers and not on formation of dimers. Biochem J 345(pt1):129–138
Sasaki T, Kikuchi T, Yumoto N, Yoshimura N, Murachi T (1984) Comparative specificity and kinetic studies on porcine calpain I and calpain II with naturally occurring peptides and synthetic fluorogenic substrates. J Biol Chem 259:12489–12494
Bussiere T, Wicinsky B, Lin GI, Perl DP, Davies P, Nixon R, Morrison JH, Hof PR (1999) Early neurodegenerative alterations in the cerebral cortex during normal aging and Alzheimer’s disease. S Neurosci Abst 25:593
Saito K, Elce JS, Hamos JE, Nixon R (1993) A. wide spread activation of calpain activated neutral proteinase (calpain) in brain in Alzheimer’s disease: A potential molecular basis for neuronal degeneration. Proc Natl Acad Sci USA 90:2628–2632
Grynspan F, Griffin WR, Cataldo A, Katayama S, Nixon RA (1997) Active site-directed antibodies identify calpain II as an early-appearing and pervasive component of neurofibrillary pathology in Alzheimer’s disease. Brain Res 763:145–158
Park SY, Ferreira A (2005) The generation of a 17 kDa neurotoxic fragment: An alternative mechanism by which tau mediates beta-amyloid-induced neurodegeneration. J Neurosci 25:5365–5375
Ferreira A, Bigio EH (2011) Calpain-mediated tau cleavage: A mechanism leading to neurodegeneration shared by multiple tauopathies. Mol Med 17:676–685
Yang J, Liu X, Bhalla K, Kim CN, Ibrado AM, Cai J, Peng I-I, Jones DP, Wang X (1997) Prevention of apoptosis by Bcl-2: Release of cytochrome c from mitochondria blocked. Science 275:1128–1132
Higaki J, Quon D, Zhong Z, Cordell B (1995) Inhibition of beta-amyloid formation identifies proteolytic precursors and subcellular site of catabolism. Neuron 14:651–659
Rechsteiner M, Rogers S, Rote K (1987) Protein-structure and intracellular stability. Trends Biochem Sci 12:390–394
Dong Y, Tan J, Cui MZ, Zhao G, Mao G, Singh N, Xu X (2005) Calpain inhibitor MDL 28170 modulates A β formation by inhibiting the formation of intermediate Aβ46 and protecting A β from degradation. FASEB J. doi:10.1096/fj.05-4524fje
Figueiredo-Pereira ME, Efthimiopoulos S, Tezapsidis N, Buku GJ, Mehta P, Robakis NK (1999) Distinct secretases, a cysteine protease and a serine protease, generate the C termini of amyloid b-proteins Ab1–40 and Ab1–42, respectively J. Neurochem 72:1417–1422
Mishizen-Eberz AJ, Guttmann RP, Giasson BI, Day GA, Hodara R, Ischiropoulos H, Lee VMY, Trojanowski JQ, Lynch DR (2003) Distinct cleavage patterns of normal and pathologic forms of a-synuclein by calpain I in vitro. J Neurochem 86:836–847
Sanchez I, Xu CJ, Juo P, Kakizaka A, Blenis J, Yuan J (1999) Caspase-8 is required for cell death induced by expanded polyglutamine repeats. Neuron 22:623–633
Hodgson JG, Agopyan N, Gutekunst CA, Leavitt BR, LePiane F et al (1999) A YAC mouse model for Huntington’s disease with full-length mutant huntingtin, cytoplasmic toxicity, and selective striatal neurodegeneration. Neuron 23(1):181–92
Paulson HL, Perez MK, Trottier Y, Trojanowski JQ, Subramony SH et al (1997) Intranuclear inclusions of expanded polyglutamine protein in spinocerebellar ataxia type 3. Neuron 19:333–344
Gould VFC, Goti D, Pearce D, Gonzalez GA, Gao H et al (2003) A Mutant Ataxin-3 Fragment results from processing at a site nterminal to amino acid 190 in brain of Machado–Joseph disease-like transgenic mice. Neurobiol Dis 27:362–369
Earnshaw WC, Martins LM, Kaufmann SH (1999) Mammalian caspases: Structure, activation, substrates, and functions during apoptosis. Annu Rev Biochem 68:383–424
Wellington CL, Ellerby LM, Gutekunst CA, Rogers D, Warby S et al (2002) Caspase cleavage of mutant huntingtin precedes neurodegeneration in Huntington’s disease. J Neurosci 22:7862–7872
Zhao M, Sua J, Heada E, Cotman CW (2003) Accumulation of caspase cleaved amyloid precursor protein represents an early neurodegenerative event in aging and in Alzheimer’s disease. Neurobiol Dis 14:391–403
Horowitz PM, Patterson KR, Guillozet-Bongaarts AL, Reynolds MR, Carroll CA et al (2004) Early N-terminal changes and caspase-6 cleavage of tau in Alzheimer’s disease. J Neurosci 24:7895–7902
Ghoshal N, García-Sierra F, Wuu J, Leurgans S, Bennett DA, Berry RW, Binder LI (2002) Tau conformational changes correspond to impairments of episodic memory in mild cognitive impairment and Alzheimer’s disease. Exp Neurol 177:475–493
Hart MJ, Glicksman M, Liu M, Sharma MK, Cuny G, Galvan V (2012) Development of a high-throughput screen targeting caspase-8-mediated cleavage of the amyloid precursor protein. Anal Biochem 421:467–476
Mejia RO, Friedlander RM (2001) Caspases in Huntington’s disease. Neuroscientist 7:480–489
Graham RK, Deng Y, Carroll J, Vaid K, Cowan C et al (2010) Cleavage at the 586 amino acid caspase-6 site in mutant huntingtin influences caspase-6 activation in vivo. J Neurosci 30:15019–15029
Zhang Y, Ona VO, Li M, Drozda M, Dubois-Dauphin M et al (2003) Sequential activation of individual caspases, and of alterations in Bcl-2 proapoptotic signals in a mouse model of Huntington’s disease. J Neurochem 87:1184–1192
Chen M, Ona VO, Li M, Ferrante RJ, Fink KB et al (2000) Minocycline inhibits caspase-1 and caspase-3 expression and delays mortality in a transgenic mouse model of Huntington disease. Nat Med 6:797–801
Wang GH, Mitsui K, Kotliarova S, Yamashita A, Nagao Y et al (1999) Caspase activation during apoptotic cell death induced by expanded polyglutamine in N2a cells. Neuroreport 10:2435–2438
Rigamonti D, Bauer JH, De-Fraja C, Conti L, Sipione S et al (2000) Wild-type huntingtin protects from apoptosis upstream of caspase-3. J Neurosci 20:3705–3713
Zhang Y, Leavitt BR, van Raamsdonk JM, Dragatsis I, Goldowitz D et al (2006) Huntingtin inhibits caspase-3 activation. EMBO J 25:5896–5906
Ona VO, Li M, Vonsattel JP, Andrews LJ, Khan SQ et al (1999) Inhibition of caspase-1 slows disease progression in a mouse model of Huntington’s disease. Nature 399:263–267
Rohn TY (2008) Caspase-Cleaved TAR DNA Binding protein-43 is a major pathological finding in Alzheimer’s disease. Brain Res 1228:189–198
Berke SJS, Schmied FAF, Brunt ER, Ellerby LM, Paulson HL (2004) Caspase-mediated proteolysis of polyglutamine disease protein ataxin-3. J Neurochem 89:908–918
Rohn TT, Kokoulina P (2009) Caspase-cleaved TAR DNA-binding protein-43 in Pick's disease. Int J Physio Pathophysio Pharmacol 20:24–31
Zhang YJ, Xu YF, Dickey CA, Buratti E, Baralle F et al (2007) Progranulin mediates caspase-dependent cleavage of TAR DNA binding protein-43. J Neurosci 27:10530–10534
Johnson BS, McCaffery M, Lindquist S, Gitler AD (2008) A yeast TDP-43 proteinopathy model: Exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc Natl Acad Sci USA 105:6439–6444
Kornfeld S (1992) Structure and function of the mannose 6-phosphate/insulin like growth factor II receptors. Ann Rev Biochem 61:307–330
Cataldo AM, Nixon RA (1990) Enzymatically active lysosomal proteases are associated with amyloid deposits in Alzheimer brain. Proc Natl Acad Sci USA 87:3861–3865
Cataldo AM, Paskevich PA, Kominami E, Nixon RA (1991) Lysosomal hydrolases of different classes are abnormally distributed in Alzheimer brain. Proc Natl Acad Sci USA 88:10998–11002
Cataldo AM, Hamilton DJ, Nixon RA (1994) Lysosomal abnormalities in degenerating neurons link neuronal compromise to senile plaque development in Alzheimer’s disease. Brain Res 640:68–80
Cataldo AM, Barnett JL, Berman SA, Quarless S, Burztajn S, Lippa C, Nixon RA (1995) Gene expression and cellular content of cathepsin D in Alzheimer’s disease brain: Evidence for early up-regulation of the endosomal–lysosomal system. Neuron 14:671–680
Jin LW, Maezawa I, Vincent I, Bird T (2004) Intracellular accumulation of amyloidogenic fragments of amyloid-β precursor protein in neurons with Niemann–Pick type C defects is associated with endosomal abnormalities. Am J Pathol 164:975–985
Matej R, Botond G, László L, Kopitar-Jerala N, Rusina R, Budka H, Kovacs GG (2010) Increased neuronal Rab5 immunoreactive endosomes do not colocalize with TDP-43 in motor neuron disease. Exp Neurol 225:133–139
Nixon RA, Cataldo AM (2006) Lysosomal system pathways: Genes to neurodegeneration in Alzheimer’s disease. J Alzheimers Dis 9:277–289
Mackay EA, Ehrhard A, Moniatte M, Guenet C, Tardif C et al (1997) A possible role for cathepsins D, E, and B in the processing of beta-amyloid precursor protein in Alzheimer’s disease. Eur J Biochem 244:414–425
Dreyer RN, Bausch KM, Fracasso P, Hammond LJ, Wunderlich D et al (1994) Processing of the pre-beta-amyloid protein by cathepsin D is enhanced by a familial Alzheimer’s disease mutation. Eur J Biochem 224:265–271
Sadik G, Kaji H, Takeda K, Yamagata F, Kameoka Y, Hashimoto K, Miyanaga K, Shinoda T (1999) In vitro processing of amyloid precursor protein by cathepsin D. Int J Biochem Cell Biol 31:1327–1337
Vigo-Pelfrey C, Lee D, Keim P, Lieberburg I, Schenk DB (1993) Characterization of beta-amyloid peptide from human cerebrospinal fluid. J Neurochem 61:1965–1968
Higaki J, Catalano R, Guzzetta AW, Quon D, Navé JF, Tarnus C, D’Orchymont H, Cordell B (1996) Processing of b-amyloid precursor protein by cathepsin D. J Biol Chem 271:31885–31893
Hook VYH, Kindy M, Hook G (2008) Inhibitors of cathepsin B improve memory and reduce β-amyloid in transgenic Alzheimer disease mice expressing the wild-type, but not the Swedish mutant, β-secretase site of the amyloid precursor protein. J Biol Chem 283:7745–7753
Vassilacopoulou D, Ripellino JA, Tezapsidis N, Hook VYH, Robakis NK (1995) Full-length and truncated APP in chromaffin granules: Solubilization of granule membrane APP by a proteolytic mechanism. J Neurochem 64:2140–2146
Hook V, Toneff T, Bogyo M, Greenbaum D, Medzihradszky KF et al (2005) Inhibition of cathepsin B reduces b-amyloid production in regulated secretory vesicles of neuronal chromaffin cells: Evidence for cathepsin B as a candidate b-secretase of Alzheimer’s disease. Biol Chem 386:931–940
Takahashi M, Ko L, Kulathingal J, Jiang P, Sevlever D, Yen SC (2007) Oxidative stress-induced phosphorylation, degradation and aggregation of α-synuclein are linked to upregulated CK2 and cathepsin D. Eur J Neurosci 26:863–874
Sevlever D, Jiang P, Yen SC (2008) Cathepsin D is the main lysosomal enzyme involved in the degradation of α-synuclein and generation of its carboxy-terminally truncated species. Biochem 47:9678–9687
Cullen V, Lindfors M, Ng J, Paetau A, Swinton E et al (2009) Cathepsin D expression level affects alpha-synuclein processing, aggregation, and toxicity in vivo. Mol Brain 2:5. doi:10.1186/1756-6606-2-5
Geiger T, Clarke S (1987) Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides. Succinimide-linked reactions that contribute to protein degradation. J Biol Chem 262:785–94
Wright HT (1991) Nonenzymatic deamidation of asparaginyl and glutaminyl residues in proteins. Crit Rev Biochem Mol Biol 26:1–52
Watanabe A, Hong WK, Dohmae N, Takio K, Morishima-Kawashima M, Ihara Y (2004) Molecular aging of tau: Disulfide-independent aggregation and non-enzymatic degradation in vitro and in vivo. J Neurochem 90:1302–1311
Watanabe A, Takio K, Ihara Y (1999) Deamidation and isoaspartate formation in smeared tau in paired helical filaments. Unusual properties of the microtubule-binding domain of tau. J Biol Chem 274:7368–7378
Dunkelberger EB, Buchanan LE, Marek P, Cao P, Raleigh DP, Zanni MT (2012) Deamidation accelerates amyloid formation and alters amylin fiber structure. J Am Chem Soc 134:12658–12667
Robinson NE, Robinson ML, Schulze SE, Lai BT, Gray HB (2009) Deamidation of alpha-synuclein. Protein Sci 18:1766–1773
Hasan Q, Alluri RV, Rao P, Ahuja YR (2006) Role of glutamine deamidation in neurodegenerative diseases associated with triplet repeat expansion – a hypothesis. J Mol Neurosci 29:29–33
Robinson NE, Robinson ZW, Robinson BR, Robinson AL, Robinson JA, Robinson ML, Robinson AB (2004) Structure-dependent nonenzymatic deamidation of glutaminyl and asparaginyl pentapeptides. J Pept Res 63:426–436
Guo H, Albrecht S, Bourdeau M, Petzke T, Bergeron C, LeBlanc AC (2004) Active caspase-6 and caspase-6-cleaved tau in neuropil threads, neuritic plaques, and neurofibrillary tangles of Alzheimer’s disease. Am J Pathol 165:523–531
Zhang Y, Xu Y, Cook C, Gendron TF, Roettges P et al (2009) Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc Natl Acad Sci USA 106:7607–7612
Acknowledgments
This work was supported by Axon Neuroscience and structural fund 26240220046.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jadhav, S., Zilka, N. & Novak, M. Protein Truncation as a Common Denominator of Human Neurodegenerative Foldopathies. Mol Neurobiol 48, 516–532 (2013). https://doi.org/10.1007/s12035-013-8440-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-013-8440-8