Abstract
Intracellular messenger RNA (mRNA) traffic and translation must be highly regulated, both temporally and spatially, within eukaryotic cells to support the complex functional partitioning. This capacity is essential in neurons because it provides a mechanism for rapid input-restricted activity-dependent protein synthesis in individual dendritic spines. While this feature is thought to be important for synaptic plasticity, the structures and mechanisms that support this capability are largely unknown. Certainly specialized RNA binding proteins and binding elements in the 3′ untranslated region (UTR) of translationally regulated mRNA are important, but the subtlety and complexity of this system suggests that an intermediate “specificity” component is also involved. Small non-coding microRNA (miRNA) are essential for CNS development and may fulfill this role by acting as the guide strand for mediating complex patterns of post-transcriptional regulation. In this review we examine post-synaptic gene regulation, mRNA trafficking and the emerging role of post-transcriptional gene silencing in synaptic plasticity.
Similar content being viewed by others
Introduction
Through its network of dendritic and axonal connections, an individual neuron may integrate information from thousands of cells. How it accomplishes this amazing feat of engineering remains a great unknown of neurobiology. What is known is that the establishment of long-term potentiation (LTP) or long-term depression (LTD) at these connections involves a combination of post-translational modification of synaptic protein and subtle changes in gene expression. While real-time changes in protein structure and function are easily reconciled, it is more difficult to imagine how a single transcriptional apparatus could respond to discrete stimuli from so many connections in a timely manner. We now know that to overcome this problem, a significant proportion of activity-associated expression is post-transcriptionally regulated in the dendritic spines of post-synaptic neurons. Understanding how this complex and dynamic temporospatial pattern can be established and encoded in functioning neurons represents a challenging biological problem. In this review, evidence and mechanisms for post-transcriptional regulation of post-synaptic gene expression are examined in the context of new information about the role of small non-coding RNA, known as microRNA.
Part I—Gene Expression and Synaptic Plasticity
In a functioning neural network, individual post-synaptic termini require the capacity to respond to direct stimuli with high specificity in real-time. While a small supply of inactive protein precursors can be maintained in the synaptic compartment, the logistics of managing activity-dependent post-translational modification of a large number of polypeptides has significant limitations. A more elegant solution is to have encoded messages present with localized translational machinery, such that peptides can be synthesized as required.
Post-synaptic Protein Synthesis
Early evidence of localized synaptic protein synthesis, the visualization of clustered ribosomes (polyribosomes) associated with the bases of dendritic spines [1], was later confirmed within isolated synaptosomes by the incorporation of radio-labeled amino acids into nascent peptides synthesized within this fraction [2]. Appearance of the labeled peptides in the synaptic membrane and post-synaptic density within 15 min of activity supported the local synthesis hypothesis, since proteins synthesized deep in the soma of a neuron could take hours to arrive at the synapse [3]. Mitochondria, themselves capable of protein synthesis, were excluded as the source of the newly synthesized peptides by differential treatment with eukaryotic and mitochondrial protein synthesis inhibitors, confirming that translation of these new peptides is indeed driven by ribosomes [4, 5].
Together these findings provide strong support for localized, on-demand synaptic protein synthesis, and to confirm that this is triggered in response to biologically relevant stimuli, Feig and Lipton demonstrated de novo dendritic protein synthesis after electrical excitation [6]. Further investigation of LTP suggested that the same signals driving axon guidance and synapse formation during development might also be involved in activity-dependent synaptic plasticity in the adult brain [7]. Supporting this hypothesis, the level of BDNF messenger RNA (mRNA) was found to increase markedly in response to electrical activity [8], evoking a proportional increase in electrical potentiation, with more rapid onset of potentiation and decreased time to elicit a 25% increase in synaptic strength [9]. This reciprocal relationship between electrical and neurotrophic signals was found to be associated with both short-term and long-term regulation of neuronal signaling (reviewed in [8]), making the neurotrophins good candidates for ongoing modulators of synaptic strength.
While these observations support the hypothesis that mRNA is actively translated into protein in dendritic spines, how is untranslated mRNA made available at the synapse in response to incoming signals?
Neuronal mRNA Traffic
Transcription, with the exception of mitochondrial encoded genes, takes place almost exclusively in the nucleus, such that synaptic mRNA must be transported large distances from this central organelle, through the dendritic tree to the dendritic spines. This poses several key logistical questions: What regulatory mechanisms are in place to ensure the correct transcripts arrive at the appropriate spines? What cellular machinery enables the mRNA to be trafficked? And importantly, how is translation suspended until the destination is reached? Detailed observation of hippocampal neurons in culture has revealed evidence of mRNA sorting even at early developmental stages (Fig. 1) [10, 11]. During the first 24 h, growing neurons sprout several minor processes in vitro that are not discernible as either axons or dendrites. The axon is the first process to extend, and its growth is rapid compared with that of dendrites. As such, polyA mRNA is found extensively at the active growth cone; there is little or no labeling of dendrites. After the axon is completed, elaboration of the dendritic tree begins. Compartmentation becomes obvious, as mRNA is no longer detectable in the axon, while labeling over the dendrites increases as the tree continues to expand.
Development of compartmentation in neurons. Neurons establish compartments very early during differentiation. Rapid growth of the axonal process results in heavy labelling of polyA mRNA over the growth cone during this period, but this is redistributed into the dendritic arbor when branching begins. In mature neurons, several protein markers can be used to identify the different types of process, demonstrating that each of the compartments has cultivated unique translational requirements
The appropriate redistribution of selective mRNA in response to specific stimuli requires an address sequence encoding the destination, and a chaperone to prevent ectopic translation en-route to the synapse. In support of this hypothesis, Steward and colleagues [12] followed the induction of mRNA for the immediate-early gene (IEG) Arc (activity-regulated cytoskeleton-associated protein) in response to synaptic activity. Arc mRNA was found to localize specifically in electrically stimulated post-synaptic dendrites, but more importantly this targeting continued when protein synthesis was abolished, suggesting the localization was signaled from within the mRNA sequence [13]. Address sequences known as cis-acting, or localization, elements (LEs) have been established for several other compartmentally enriched mRNA by observing disrupted localization resulting from specific deletions or antisense molecules. A 54-nucleotide conserved element in the 3′-untranslated region (UTR) of the β-actin transcript was found to direct the localization of the mRNA to growth cones of cultured neurons in response to neurotransmitter (NT-3) stimulation. With anti-sense inhibition of the zipcode sequence, the neurotransmitter still elicited an increase in β-actin mRNA synthesis; however, this RNA interference prevented the formation of a trans-acting ribonucleoprotein (RNP) complex with Zipcode Binding Protein-1 (ZBP1), and the transcript was confined to the soma [14].
Similar cis-acting elements have been reported in the 3′-UTRs of other spatially regulated transcripts including myelin basic protein (MBP) in oligodendrocytes [15], elongation factor 1α [16], microtuble-associated protein 2 (MAP2) [17], and the α-subunit of calmodulin-dependent protein kinase 2 (CaMKIIα) [18]. In the latter two cases, however, the targeting element identified is not the sole determinant of localization, suggesting the existence of a more complex system for regulating the expression of genes after transcription. LEs of 640 and 94 nucleotides in the 3′-UTRs of MAP2 and CaMKIIα, respectively, were found to be sufficient for dendritic localization. In the case of MAP2, cells expressing a 3′-UTR with the LE deleted exhibited slightly reduced targeting compared with those expressing shorter partial UTRs, which the authors proposed could arise from alteration of the UTR’s secondary structure after specific deletion of the LE [17]. Furthermore, while the 94-nt cis-acting element found in CaMKIIα successfully targeted the mRNA to dendrites, partial deletions of this region, which retained the LE, failed to localize. However, targeting of this construct was restored by depolarization with BDNF, implying the involvement of a second, inhibitory cis-acting element that functions to suppress transport in resting cells [18]. Microarray analysis of mRNA differentially associated with polysomes after BDNF stimulation agreed with this finding and identified several other candidates for dendritic targeting and translation, including N-methyl d-aspartate (NMDA) receptor subunit 3 (NR3), and the PSD scaffolding protein Homer2, which were validated in preparations of rat forebrain synaptoneurosomes [19].
Although the mechanisms are different, findings regarding CaMKII and MAP2 are suggestive of external interference acting upon the 3′-UTRs of dendritically active mRNA. One method by which the secondary structure of MAP2 could be altered is by the reversible binding of silencing protein or RNA entities within the LE, which could be removed in response to activity and restore functionality. This same strategy could explain the activity-induced de-repression exhibited by the secondary cis-acting element of the CaMKIIα UTR. Moreover, it is interesting to note that both mRNA were co-localized with distinct moving granules. Together these observations suggest a system of reversible suppression, perhaps utilizing the RNA interference (RNAi) pathway and post-transcriptional gene silencing (PTGS).
Part II—Post-transcriptional Gene Silencing
Within the last 10 years, the discovery of endogenous small non-coding RNA known as microRNA (miRNA) has sparked a flurry of research and dramatically changed our understanding of gene regulation. miRNA employ complimentary base pairing and the RNA induced silencing complex (RISC) to bind and either suppress translation or facilitate degradation of their target mRNA. A large proportion of these molecules are enriched or specifically expressed in the mammalian brain, and many of these also co-purify in neurons with actively translating polyribosomes [20], thereby potentially regulating protein expression at the synapse in response to changing requirements.
MicroRNA
miRNA are 19–22-nucleotide RNA fragments derived primarily from non-coding regions of the genome, although some are embedded within the coding regions of known genes [21]. miRNA are transcribed into long primary transcripts (pri-miRNA) forming hairpin structures that are cleaved in the nucleus by the microprocessor complex consisting primarily of Drosha and DGCR8, to form shorter (~ 70 nt) precursor hairpins known as pre-miRNA (Fig. 2). Exportin-5 transfers pre-miRNA to the cytoplasm for further cleavage by Dicer to yield the mature, double-stranded miRNA (~ 20 nt). The mature miRNA becomes active after it is assembled into the RISC with proteins from the Argonaute (Ago) family. After selective removal and degradation of the passenger strand [22–24], the RISC-associated guide strand directs the complex in search of its mRNA targets.
miRNA biogenesis pathway. Primary miRNA (pri-miR) are transcribed from genomic DNA in the nucleus where they are cleaved by Drosha, complexed with DGCR8, to shorter precursor transcripts (pre-miR) ~ 80–100 nucleotides in length. Exportin-5 transports pre-miRs to the cytoplasm where Dicer cleaves them to a duplex structure (~ 20 nucleotides) that is loaded into the RISC. The passenger strand is ejected from the RISC and degraded, while the guide strand identifies the mRNA target and binds it, resulting in deadenylation and degradation, or translational repression, of the transcript
The base pairing of miRNA and mRNA in vertebrates only requires partial homology, with a preference for contiguous pairing only occurring at the “seed” region, located at nucleotides two through seven of the guide strand. The lack of stringency results in a many-to-many relationship between miRNA and mRNA targets, with the consequence that as much as 60% of the genome may be regulated post-transcriptionally by a comparatively small cohort of miRNA [23]. The guide strand binds to the complimentary region in the 3′ UTR of its target by Watson–Crick base pairing of the seed residues. Several alternative seed binding arrangements have been observed (Fig. 3), and these are believed to confer differential target affinity, proportional to the number of nucleotides involved [25]. There is no specific binding between the Ago domains and the miRNA; rather the strand is held in place by energetically favorable pi-stacking bonds between the aromatic rings of the nucleotides and conserved phenylalanine residues in the binding pocket [26], suggesting that if the RISC were disassembled its cargo would be freed without modification. This raises the possibility of miRNA recycling since some enzymes employ this method of engagement to participate transiently in catalytic reactions. The potential for de-repression suggests a possible role for miRNA in reversible and cyclic transcript silencing.
miRNA-target binding. Canonical binding (a–c) includes the seed residues (2–7) and either (a, b) or both (c) residues 1 and 8. Marginal binding is only 6-mer, involving only the seed residues (d) or an offset including residue 8 (e). Canonical binding is considered stronger and more effective for target recognition compared with marginal binding. (Adapted from [25])
miRNA in Neural Structure and Function
A key role for processed miRNA in neuronal differentiation is demonstrated by significantly increased Dicer expression in response to differentiation in both neuroblastoma cell lines [27] and cultures of cerebellar granule neurons [28]. These models have identified miRNA that are both up-regulated and down-regulated by the differentiation process. Importantly, enhanced expression of known brain-specific and brain-enriched miRNA including miR-124a was reported, while down-regulation of the miR-17 family, whose targets are enriched for pathways involving LTP, axon guidance and regulation of the actin cytoskeleton [29], is consistent with the expectation that these mRNA would be in high demand during this period. Further, miR-125b also exhibited up-regulation during differentiation and was shown to be both necessary and sufficient for neurite outgrowth in the differentiating cells by both gain- and loss-of-function studies [30].
The impact of miRNA on development is exemplified in studies where miRNA maturation was ablated by Dicer knock-out [31, 32]. Although miRNA depletion appears to have little effect on uncommitted progenitor cells, which retain their ability to proliferate [32], differentiation and morphogenesis are clearly affected. The resulting phenotypes are grossly abnormal, exhibiting improper differentiation, incomplete neural patterning including reduced arealization and layering, lack of interneurons, and impaired connectivity, dendritic targeting and arborization [31–33], suggesting that miRNA assist in providing a biologically robust environment during development [34, 35]. With an emerging role in neural development, it is not surprising that alterations in miRNA biogenesis and expression are also associated with neurodevelopmental disorders such as schizophrenia [29, 36–38].
Arguably one of the most studied CNS-specific miRNA is miR-124, which has been shown to regulate the switch from progenitor cell to committed neuron, acting in one instance through Sox9 to switch off glial function and promote cell cycle exit leading to activation of the differentiation process [39]. Along with miR-9*, miR-124 is suppressed in neural progenitors by the repressor-element-1-silencing transcription factor (REST). When differentiation is initiated, REST suppression is lifted, and the miRNA targets the 3′-UTR of chromatin remodeling factor BAF53a, thereby epigenetically mediating the switch to a differentiated pattern of gene expression [40]. Interestingly, ectopic expression of miR-124 in association with dFMR1, the homolog of fragile-X mental retardation protein (FMRP) in Drosophila, decreased dendritic branching in dendritic arborization sensory neurons [41].
This intricacy and diversity of miR-124’s activities during neural development underscores several key points regarding the current understanding of miRNA biology. Firstly, while many brain-specific and brain-enriched miRNA are critical for correct development of the brain and nervous system, very few have been characterized in detail. Secondly, neural miRNA have many targets and participate in a range of complexes to achieve their regulatory goals. Thirdly, and perhaps most importantly, miRNA can promote the neuronal phenotype while at the same time negatively regulating some neural functions, such as miR-124 and dendritic branching. This is to be expected as the influence of miRNA is a continuum that “fine-tunes” mRNA expression rather than switching it on and off, where the balance of activity may vary spatially within the cell at any given time.
Part III—Could miRNA and PTGS Provide a Mechanism for Activity-dependent mRNA Translation at the Synapse?
Remodeling of discreet post-synaptic membranes in response to activity requires mechanisms that facilitate real-time control of protein synthesis. An important aspect of this control is defined by the local availability and activity of mRNA. While this is supported by a plethora of sequence specific solutions, the evolution of nucleic acid-dependent gene silencing pathways has also provided neurons with an opportunity to employ a more universal system for achieving these objectives.
The key advantage of using nucleic acids over proteins to implement cellular trafficking is the flexibility provided by the level of functional redundancy built into the system. Using individual carrier proteins to chaperone each transcript around the cell is a fairly limited solution, which becomes a very difficult problem in complex systems. By utilizing an RNA-encoded adapter molecule such as miRNA, the post-transcriptional regulatory systems are provided with the potential for extraordinary autonomy, economy and scalability that may enable them to manage the complexity required for sophisticated neural systems.
miRNA and Post-synaptic Gene Regulation
Studies of miRNA expression in synaptoneurosomes suggest enrichment and depletion of subsets of miRNA in the vicinity of synapses [42, 43], where they have demonstrated key roles regulating spine morphology and excitability [43–47], briefly summarized in Table 1. Pre-miRNA and an inactive form of Dicer have additionally been observed localized to the PSD [42, 48]. This is significant in the context of synaptic function because increased intracellular Ca2+ driven by NMDA stimulation has been shown to induce calpain-directed cleavage and activation of Dicer, which can subsequently process pre-miRNA into functionally active mature miRNA.
Conversely, excitation may also lift the repression imposed on synaptically localized RISC-associated mRNA. In support of this hypothesis the RISC protein Armitage has been shown to decompose in response to LTP-inducing stimuli in Drosophila neurons, releasing CaMKII mRNA and other memory-associated transcripts for immediate translation [49]. While the CaMKII 3′-UTR was shown to be regulated by miR-280 and miR-289, it seems unlikely that these are the only miRNA regulating this transcript. Like many dendritically localized mRNA such as MAP2, CaMKIIα has a long 3′-UTR with many predicted miRNA binding sites. The secondary structure of the MAP2 3′-UTR was proposed to be responsible for its dendritic localization [17], and based on understanding of miRNA function, it is possible that miRNA bind to the 3′ UTR and transport MAP2, and potentially other plasticity-associated mRNA, to the dendrites. Interestingly, many of these transcripts co-localize with neuronal granules. These ribonucleoprotein complexes are associated with synapto-dendritic transport and trafficking. As these structures also contain miRNA, they are likely to play a role in post-synaptic gene silencing.
Neuronal Granules
Several classes of neuronal granule have been shown to co-localize with miRNA, mRNA and other known protein and enzyme components of RNAi (reviewed in [50]). Granules were originally discovered as GW bodies and are marked by the RNA-binding protein GW182. Their primary role appears to be the spatial control of mRNA availability via 5′-3′ mediated decay, and they are associated with various mRNA-degrading enzymes. GW bodies are now more commonly referred to as processing bodies (P-bodies) and are known in many cell types as post-transcriptional regulators of gene expression [51], chiefly facilitating pathways such as nonsense mediated decay, RNA degradation and miRNA-mediated silencing [52]. Another major group of granule detected in the cytoplasm of neurons is transport RNPs (tRNPs) whose markers include Staufen1 and 2, FMRP and ZBP1, and which are chiefly involved in redistribution of translationally arrested mRNA [53]. A group of heterogeneous nuclear RNPs (hnRNPs) is also active in the nucleus. These nuclear granules interact with their cytoplasmic counterparts via nuclear export factor 7 (NXF7) to coordinate mRNA sorting, transport and storage [54].
The heterogeneity of neuronal granules and relative paucity of known markers has led to conflict in their elaboration, with some studies reporting tRNPs to be similar in structure and function to P-bodies [55], while others report them distinct [56]. Some of this confusion might be explained by reports of transient physical association between P-bodies and stress granules [57], and more recent video-microscopic reports of docking events between tRNPs and P-bodies [56]. These have been observed with varying time-courses and could certainly confound observations depending on the labeling and timing protocols employed. The addition of Ago-containing miRNA-induced RNPs (miRNPs) further supports a model involving diverse, functionally distinct granules. Not only are the Ago proteins themselves functionally distinct, but expression profiling of miRNA obtained after incubation with Ago1-specific antibodies selected target mRNA that were enriched for long 3′-UTRs, demonstrating a selection process in the loading of targets into Ago1 miRNPs, the significance of which is not yet understood [58].
A neuron-specific sub-class of granule known as dlP-bodies has recently been identified and also contains ribosomes and enzymes required for localized protein translation, while being depleted of XRN1 normally associated with P-bodies that induce RNA decay [59]. DlP-bodies exhibit directed movement towards distant synapses in response to BDNF stimulation where they subsequently disassemble, freeing the mRNA for potential translation, and indeed neurons have been demonstrated to increase localized mRNA translation at the synapse in response to BDNF. Similar results have been observed in other sub-types of P-body in response to NMDA and glutamate [56], with the different depolarization agents shown to affect overall composition of P-body and tRNP subpopulations. Together these findings suggest that neuronal granules play an important role in miRNA-associated dendrito-synaptic trafficking of mRNA, as well as real-time, post-synaptic, activity-dependent regulation of translation in response to incoming signals.
Functional Integration
The evidence discussed strongly supports the existence of a deliberate, highly sophisticated and coordinated system of mRNA sorting, transportation and localization in mammalian neurons. To ensure the smooth running of such a network undoubtedly requires extensive inter-compartmental communication. The nature of much of this signaling remains undetermined; however, as a common participant in all compartments, miRNA undoubtedly play a key role. The recent detection of docking events between tRNPs and P-bodies provides an interesting mechanism by which communication could be facilitated. Docking was observed over periods up to 1 min, during which time it is conceivable that mRNA could be “rescued” from degradation by transferring from P-body to tRNP; conversely, transcripts that are no longer required may be moved from tRNP to P-body for disposal. Alternatively, it may be that the different classes of granules dock on some occasions to exchange information “updates” regarding the transcriptional requirements of the cell. Given that new classes of granule continue to be identified, the protein markers of the various classes remain largely undefined, and the contents (and therefore potential functions) differ greatly between granules, this area of research is likely to yield interesting insights into intracellular communication.
Could miRNA Compartmentalization Regulate Activity-driven Synaptic Gene Expression and Memory Formation?
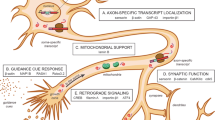
In this review we have drawn on the current evidence that supports an integrated model of the synaptic response to excitation (Fig. 4). In the resting state (Fig. 4, left panel), translationally repressed transcripts are localized near the bases of dendritic spines, along with ribosomes, while inactive Dicer and pre-miRNA are anchored in the PSD. In response to stimulation by neurotransmitters or neurotrophic factors, several cascades of activity are initiated in the post-synaptic cell (Fig. 4, center panel). The rapid increase in intracellular calcium activates calpain, which cleaves Dicer from the PSD; pre-miRNA are also released, although the mechanism driving this is unknown. Stimulation causes mRNA-associated RISCs to decompile, allowing the transcripts to be translated by polyribosomes. As depolarization continues (Fig. 4, right panel), active Dicer is deployed, cleaving pre-miRNA to provide more mature miRNA to perhaps sequester free translating mRNA and provide some attenuation to the stimulus pulse.
Involvement of PTGS in synaptic plasticity. In the resting synapse (left panel), the key role of PTGS is the silencing of mRNA functionally localized here but not yet required for translation; however, inactive elements of the miRNA pathway may also be present and tethered in the PSD, such as Dicer and pre-miRNA. When a stimulus arrives (center panel), the increase in calcium (Ca2+) triggers events that alter the translational equilibrium of the synapse. These may include de-repression and translation of RISC-bound mRNA, activation of Dicer and release of pre-miRNA. As depolarisation continues (right panel), Dicer may cleave new mature miRNA to counterbalance freed mRNA as the new equilibrium of the synapse is realized
It should be emphasized that not all these events need occur in response to a discrete excitation. Rather, these mechanisms are more likely employed as needed to modulate what can be regarded as the translational homeostasis of each individual synapse, and this will necessarily vary between synapses depending on the number and nature of their connections. The participation of miRNA and other elements of post-transcriptional gene silencing in so many aspects of the synaptic response to excitation indicates that these small regulatory molecules may be key modulators of mRNA availability and protein synthesis in post-synaptic termini. Some aspects of this integrated model have been demonstrated in response to patterns of stimulation normally associated with the formation of long-term and/or short- term memory; however, of almost 1000 known human miRNA, very few have begun to be characterized, with many studies restricting their focus to targets of individual miRNA or related family members. In this respect, broader studies of the subcellular redirection of both mRNA and miRNA in response to physiological stimuli may prove more informative in the full elaboration of the contribution of reversible gene silencing to synaptic plasticity in the context of functioning neurons and the pathophysiology of neurological disorders.
References
Steward O, Levy WB (1982) Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J Neurosci 2(3):284–291
Rao A, Steward O (1991) Evidence that protein constituents of postsynaptic membrane specializations are locally synthesized: analysis of proteins synthesized within synaptosomes. J Neurosci 11(9):2881–2895
Steward O, Pollack A, Rao A (1991) Evidence that protein constituents of postsynaptic membrane specializations are locally synthesized: time course of appearance of recently synthesized proteins in synaptic junctions. J Neurosci Res 30(4):649–660
Torre ER, Steward O (1992) Demonstration of local protein synthesis within dendrites using a new cell culture system that permits the isolation of living axons and dendrites from their cell bodies. J Neurosci 12(3):762–772
Kang H, Schuman EM (1996) A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science 273(5280):1402–1406
Feig S, Lipton P (1993) Pairing the cholinergic agonist carbachol with patterned Schaffer collateral stimulation initiates protein synthesis in hippocampal CA1 pyramidal cell dendrites via a muscarinic, NMDA-dependent mechanism. J Neurosci 13(3):1010–1021
Goodman CS, Shatz CJ (1993) Developmental mechanisms that generate precise patterns of neuronal connectivity. Cell 72(Suppl):77–98
Lo DC (1995) Neurotrophic factors and synaptic plasticity. Neuron 15(5):979–981
Kang H et al (1996) Determinants of BDNF-induced hippocampal synaptic plasticity: role of the Trk B receptor and the kinetics of neurotrophin delivery. Learn Mem 3(2–3):188–196
Dotti CG, Sullivan CA, Banker GA (1988) The establishment of polarity by hippocampal neurons in culture. J Neurosci 8(4):1454–1468
Kleiman R, Banker G, Steward O (1994) Development of subcellular mRNA compartmentation in hippocampal neurons in culture. J Neurosci 14(3 Pt 1):1130–1140
Steward O et al (1998) Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron 21(4):741–751
Dahm R, Kiebler M, Macchi P (2007) RNA localisation in the nervous system. Semin Cell Dev Biol 18(2):216–223
Zhang HL et al (2001) Neurotrophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron 31(2):261–275
Ainger K et al (1997) Transport and localization elements in myelin basic protein mRNA. J Cell Biol 138(5):1077–1087
Huang F, Chotiner JK, Steward O (2005) The mRNA for elongation factor 1alpha is localized in dendrites and translated in response to treatments that induce long-term depression. J Neurosci 25(31):7199–7209
Blichenberg A et al (1999) Identification of a cis-acting dendritic targeting element in MAP2 mRNAs. J Neurosci 19(20):8818–8829
Mori Y et al (2000) Two cis-acting elements in the 3′ untranslated region of alpha-CaMKII regulate its dendritic targeting. Nat Neurosci 3(11):1079–1084
Schratt GM et al (2004) BDNF regulates the translation of a select group of mRNAs by a mammalian target of rapamycin-phosphatidylinositol 3-kinase-dependent pathway during neuronal development. J Neurosci 24(33):7366–7377
Kim J et al (2004) Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci USA 101(1):360–365
Zhu Y et al (2009) A microRNA gene is hosted in an intron of a schizophrenia-susceptibility gene. Schizophr Res 109(1–3):86–89
Bicker S, Schratt G (2008) MicroRNAs: tiny regulators of synapse function in development and disease. J Cell Mol Med 12(5A):1466–1476
Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9(2):102–114
Konecna A, et al. 2009 What are the roles of microRNAs at the mammalian synapse? Neurosci Lett
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136(2):215–233
Jinek M, Doudna JA (2009) A three-dimensional view of the molecular machinery of RNA interference. Nature 457(7228):405–412
Potenza N, Papa U, Russo A (2009) Differential expression of Dicer and Argonaute genes during the differentiation of human neuroblastoma cells. Cell Biol Int 33(7):734–738
Barbato C et al (2007) Dicer expression and localization in post-mitotic neurons. Brain Res 1175:17–27
Beveridge NJ, et al (2009) Down-regulation of miR-17 family expression in response to retinoic acid induced neuronal differentiation. Cell Signal 21(12):1837–1845
Le MTN et al (2009) MicroRNA-125b promotes neuronal differentiation in human cells by repressing multiple targets. Mol Cell Biol 29(19):5290–5305
Giraldez AJ et al (2005) MicroRNAs regulate brain morphogenesis in zebrafish. Science 308(5723):833–838
De Pietri Tonelli D et al (2008) miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development 135(23):3911–3921
Berdnik D et al (2008) MicroRNA processing pathway regulates olfactory neuron morphogenesis. Curr Biol 18(22):1754–1759
Li X et al (2009) A microRNA imparts robustness against environmental fluctuation during development. Cell 137(2):273–282
Stark A et al (2005) Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell 123(6):1133–1146
Beveridge NJ et al (2010) Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol Psychiatry 15(12):1176–1189
Gardiner E, et al. 2011 Imprinted DLK1-DIO3 region of 14q32 defines a schizophrenia-associated miRNA signature in peripheral blood mononuclear cells. Mol Psychiatry. (July 5, EPUB ahead of print)
Santarelli DM et al (2011) Upregulation of dicer and microRNA expression in the dorsolateral prefrontal cortex Brodmann area 46 in schizophrenia. Biol Psychiatry 69(2):180–187
Cheng L-C et al (2009) miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci 12(4):399–408
Yoo A (2009) et al. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development, Nature
Xu X-L et al (2008) The steady-state level of the nervous-system-specific microRNA-124a is regulated by dFMR1 in Drosophila. J Neurosci 28(46):11883–11889
Lugli G et al (2008) Expression of microRNAs and their precursors in synaptic fractions of adult mouse forebrain. J Neurochem 106(2):650–661
Siegel G et al (2009) A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol 11(6):705–716
Edbauer D et al (2010) Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 65(3):373–384
Fiore R et al (2009) Mef2-mediated transcription of the miR379-410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J 28(6):697–710
Rajasethupathy P et al (2009) Characterization of small RNAs in aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron 63(6):803–817
Schratt GM et al (2006) A brain-specific microRNA regulates dendritic spine development. Nature 439(7074):283–289
Lugli G et al (2005) Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J Neurochem 94(4):896–905
Ashraf SI et al (2006) Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell 124(1):191–205
Eulalio A, Behm-Ansmant I, Izaurralde E (2007) P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol 8(1):9–22
Jakymiw A et al (2007) The role of GW/P-bodies in RNA processing and silencing. J Cell Sci 120(Pt 8):1317–1323
Eulalio A et al (2007) P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol 27(11):3970–3981
Kiebler MA, Bassell GJ (2006) Neuronal RNA granules: movers and makers. Neuron 51(6):685–690
Katahira J et al (2008) Nuclear RNA export factor 7 is localized in processing bodies and neuronal RNA granules through interactions with shuttling hnRNPs. Nucleic Acids Res 36(2):616–628
Barbee SA et al (2006) Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron 52(6):997–1009
Zeitelhofer M et al (2008) Dynamic interaction between P-bodies and transport ribonucleoprotein particles in dendrites of mature hippocampal neurons. J Neurosci 28(30):7555–7562
Kedersha N et al (2005) Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol 169(6):871–884
Hong X, et al. 2009 Immunopurification of Ago1 miRNPs selects for a distinct class of microRNA targets. Proc Natl Acad Sci USA.
Cougot N et al (2008) Dendrites of mammalian neurons contain specialized P-body-like structures that respond to neuronal activation. J Neurosci 28(51):13793–13804
Acknowledgments
MJC is supported by the Schizophrenia Research Institute and an M.C. Ainsworth Research Fellowship in Epigenetics. BJG is supported by an APA scholarship. MJC’s research is supported by the Schizophrenia Research Institute; a NARSAD Young Investigator Award; a Hunter Medical Research Institute project grant; an NHMRC project grant (631057) and the Neurobehavioral Genetics Unit, utilising funding from NSW Health.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Goldie, B.J., Cairns, M.J. Post-Transcriptional Trafficking and Regulation of Neuronal Gene Expression. Mol Neurobiol 45, 99–108 (2012). https://doi.org/10.1007/s12035-011-8222-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-011-8222-0