Abstract
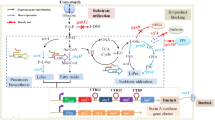
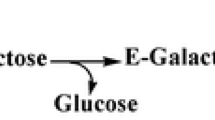
2′-Fucosyllactose (2′-FL), one of the most abundant oligosaccharides in human milk, has gained increased attention owing to its nutraceutical and pharmaceutical potential. However, limited availability and high-cost of preparation have limited its widespread application and in-depth investigation of its potential functions. Here, a modular pathway engineering was implemented to construct an Escherichia coli strain to improve the biosynthesis titer of 2′-FL. Before overexpression of manB, manC, gmd, wcaG, and heterologous expression of futC, genes wcaJ and lacZ encoding UDP-glucose lipid carrier transferase and β-galactosidase, respectively, were inactivated from E. coli BL21 (DE3) with the CRISPR-Cas9 system, which inhibited the production of 2′-FL. The results showed that final shake flask culture yielded a 3.8-fold increase in 2′-FL (0.98 g/L) from the engineered strain ELC07. Fed-batch fermentation conditions were optimized in a 3-L bioreactor. The highest titer of 2′-FL (18.22 g/L) was obtained, corresponding to a yield of 0.25 g/g glycerol and a substrate conversion of 0.88 g/g lactose.
Graphical Abstract








Similar content being viewed by others
Data Availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Li, M., Li, C., Hu, M., & Zhang, T. (2022). Metabolic engineering strategies of de novo pathway for enhancing 2′-fucosyllactose synthesis in Escherichia coli. Microbial Biotechnology, 15, 1561–1573.
Bode, L. (2012). Human milk oligosaccharides: Every baby needs a sugar mama. Glycobiology, 22(9), 1147–1162.
Thongaram, T., Hoeflinger, J. L., Chow, J., & Miller, M. J. (2017). Human milk oligosaccharide consumption by probiotic and human-associated Bifidobacteria and Lactobacilli. Journal of Dairy Science, 100(10), 7825–7833.
Vandenplas, Y., Berger, B., Carnielli, V. P., Ksiazyk, J., Lagström, H., Sanchez-Luna, M., Migacheva, N., Mosselmans, J. M., Picaud, J. C., Possner, M., Singhal, A., & Wabitsch, M. (2018). Human milk oligosaccharides: 2′-fucosyllactose (2′-FL) and lacto-N-neotetraose (LNnT) in infant formula. Nutrients, 10(9), 1161.
Bienenstock, J., Buck, R. H., Linke, H., Forsythe, P., Stanisz, A. M., & Kunze, W. A. (2013). Fucosylated but not sialylated milk oligosaccharides diminish colon motor contractions. PLoS ONE, 8(10), e76236.
Jacobi, S. K., Yatsunenko, T., Li, D., Dasgupta, S., Yu, R. K., Berg, B. M., Chichlowski, M., & Odle, J. (2016). Dietary isomers of sialyllactose increase ganglioside sialic acid concentrations in the corpus callosum and cerebellum and modulate the colonic microbiota of formula-fed piglets. Journal of Nutrition, 146(2), 200–208.
Lee, W. H., Pathanibul, P., Quarterman, J., Jo, J. H., Han, N. S., Miller, M., Jin, Y. S., & Seo, J. H. (2012). Whole cell biosynthesis of a functional oligosaccharide, 2′-fucosyllactose, using engineered Escherichia coli. Microbial Cell Factories, 11, 48.
Hegar, B., Wibowo, Y., Basrowi, R. W., Ranuh, R. G., Sudarmo, S. M., Munasir, Z., Atthiyah, A. F., Widodo, A. D., Supriatmo, K. M., Suryawan, A., Diana, N. R., Manoppo, C., & Vandenplas, Y. (2019). The role of two human milk oligosaccharides, 2′-fucosyllactose and lacto-N-neotetraose, in infant nutrition. Pediatric Gastroenterology, Hepatology & Nutrition, 22(4), 330–340.
Castanys-Muñoz, E., Martin, M. J., & Prieto, P. A. (2013). 2′-Fucosyllactose: An abundant, genetically determined soluble glycan present in human milk. Nutrition Reviews, 71(12), 773–789.
Li, W., Zhu, Y., Wan, L., Guang, C., & Mu, W. (2021). Pathway optimization of 2′-fucosyllactose production in engineered Escherichia coli. Journal of Agricultural and Food Chemistry, 69(5), 1567–1577.
Baumgärtner, F., Seitz, L., Sprenger, G. A., & Albermann, C. (2013). Construction of Escherichia coli strains with chromosomally integrated expression cassettes for the synthesis of 2′-fucosyllactose. Microbial Cell Factories, 12, 1–13.
Chin, Y. W., Park, J. B., Park, Y. C., Kim, K. H., & Seo, J. H. (2013). Metabolic engineering of Corynebacterium glutamicum to produce GDP-l-fucose from glucose and mannose. Bioprocess and Biosystems Engineering, 36(6), 749–756.
Huang, D., Yang, K., Liu, J., Xu, Y., Wang, Y., Wang, R., Liu, B., & Feng, L. (2017). Metabolic engineering of Escherichia coli for the production of 2′-fucosyllactose and 3-fucosyllactose through modular pathway enhancement. Metabolic Engineering, 41, 23–38.
Ishihara, H., & Heath, E. C. (1968). The metabolism of l-fucose IV. The biosyntthesis of guanosine diphosphate l-fucose in porcine liver. Journal of Biological Chemistry, 243, 1110–1115.
Park, S. H., Pastuszak, I., Drake, R., & Elbein, A. D. (1998). Purification to apparent homogeneity and properties of pig kidney l-fucose kinase. Journal of Biological Chemistry, 273, 5685–5691.
Pastuszak, I., Ketchum, C., Hermanson, G., Sjöberg, E. J., Drake, R., & Elbein, A. D. (1998). GDP-l-fucose pyrophosphorylase purification, cDNA cloning, and properties of the enzyme. Journal of Biological Chemistry, 273, 30165–30174.
Liu, T. W., Ito, H., Chiba, Y., Kubota, T., Sato, T., & Narimatsu, H. (2011). Functional expression of l-fucokinase/guanosine 5′-diphosphate-l-fucose pyrophosphorylase from Bacteroides fragilis in Saccharomyces cerevisiae for the production of nucleotide sugars from exogenous monosaccharides. Glycobiology, 21(9), 1228–1236.
Michael, J., Coyne, B. R., Martin, M. L., & Laurie, E. C. (2005). Human symbionts use a host-like pathway for surface fucosylation. Science, 307, 1778–1781.
Albermann, C., Piepersberg, W., & Wehmeier, U. F. (2001). Synthesis of the milk oligosaccharide 2′-fucosyllactose using recombinant bacterial enzymes. Carbohydrate Research, 334, 97–103.
Chin, Y. W., Seo, N., Kim, J. H., & Seo, J. H. (2016). Metabolic engineering of Escherichia coli to produce 2′-fucosyllactose via salvage pathway of guanosine 5′-diphosphate (GDP)-l-fucose. Biotechnology and Bioengineering, 113(11), 2443–2452.
Chin, Y. W., Kim, J. Y., Lee, W. H., & Seo, J. H. (2015). Enhanced production of 2′-fucosyllactose in engineered Escherichia coli BL21star (DE3) by modulation of lactose metabolism and fucosyltransferase. Journal of Biotechnology, 210, 107–115.
Jiang, W., Bikard, D., Cox, D., Zhang, F., & Marraffini, L. A. (2013). RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nature Biotechnology, 31(3), 233–239.
Jiang, Y., Chen, B., Duan, C., Sun, B., Yang, J., & Yang, S. (2015). Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Applied and Environmental Microbiology, 81(7), 2506–2514.
Zhao, D., Yuan, S., Xiong, B., Sun, H., Ye, L., Li, J., Zhang, X., & Bi, C. (2016). Development of a fast and easy method for Escherichia coli genome editing with CRISPR/Cas9. Microbial Cell Factories, 15(1), 205.
Zerbini, F., Zanella, I., Fraccascia, D., König, E., Irene, C., Frattini, L. F., Tomasi, M., Fantappiè, L., Ganfini, L., Caproni, E., Parri, M., Grandi, A., & Grandi, G. (2017). Large scale validation of an efficient CRISPR/Cas-based multi gene editing protocol in Escherichia coli. Microbial Cell Factories, 16(1), 68.
Fordjour, E., Adipah, F. K., Zhou, S., Du, G., & Zhou, J. (2019). Metabolic engineering of Escherichia coli BL21 (DE3) for de novo production of l-DOPA from d-glucose. Microbial Cell Factories, 18(1), 74.
Zhao, M., Huang, D., Zhang, X., Koffas, M. A. G., Zhou, J., & Deng, Y. (2018). Metabolic engineering of Escherichia coli for producing adipic acid through the reverse adipate-degradation pathway. Metabolic Engineering, 47, 254–262.
Di, H., Yang, K., Liu, J., Xu, Y., Wang, Y., Wang, R., Liu, B., & Feng, L. (2017). Metabolic engineering of Escherichia coli for the production of 2′-fucosyllactose and 3-fucosyllactose through modular pathway enhancement. Metabolic Engineering, 41, 23–38.
Wan, L., Zhu, Y., Li, W., Zhang, W., & Mu, W. (2020). Combinatorial modular pathway engineering for guanosine 5′-diphosphate-l-fucose production in recombinant Escherichia coli. Journal of Agricultural and Food Chemistry, 68(20), 5668–5675.
Korz, D. J., Rinas, U., Hellmuth, K., Sanders, E. A., & Deckwer, W. D. (1995). Simple fed-batch technique for high cell density cultivation of Escherichia coli. Journal of Biotechnology, 39, 59–65.
Young, L. I., & Sang, Y. L. (1996). Enhanced production of poly(3-hydroxybutyrate) by filamentation-suppressed recombinant Escherichia coli in a defined medium. Journal of Polymers and the Environment, 4, 131–134.
Wu, H., Chen, S., Ji, M., Chen, Q., Shi, J., & Sun, J. (2019). Activation of colanic acid biosynthesis linked to heterologous expression of the polyhydroxybutyrate pathway in Escherichia coli. International Journal of Biological Macromolecules, 128, 752–760.
Sander, J. D., & Joung, J. K. (2014). CRISPR-cas systems for editing, regulating and targeting genomes. Nature Biotechnology, 32(4), 347–355.
Faijes, M., Castejón-Vilatersana, M., Val-Cid, C., & Planas, A. (2019). Enzymatic and cell factory approaches to the production of human milk oligosaccharides. Biotechnology Advances, 37(5), 667–697.
Puertas, J. M., Ruiz, J., Vega, M. R. D. L., Lorenzo, J., Caminal, G., & González, G. (2010). Influence of specific growth rate over the secretory expression of recombinant potato carboxypeptidase inhibitor in fed-batch cultures of Escherichia coli. Process Biochemistry, 45(8), 1334–1341.
Funding
This work was financially supported by Major Science and Technology Innovation Project of Shandong Province (Grant No. 2020CXGC010601) and Kitty Hawk Project of Zhejiang Provincial Administration for Market Regulation (Grant No. CY2022004).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by CL and ML. The first draft of the manuscript was written by CL. Reviewing and editing of the manuscript were performed by ML and MH. Supervision, giving major comments, and project administration were contributed by TZ. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors have no relevant financial or non-financial interests to disclose.
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Consent to Publish
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, C., Li, M., Hu, M. et al. Metabolic Engineering of De Novo Pathway for the Production of 2′-Fucosyllactose in Escherichia coli. Mol Biotechnol 65, 1485–1497 (2023). https://doi.org/10.1007/s12033-023-00657-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-023-00657-7