Abstract
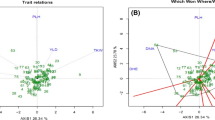
The main goals of the present study were to screen Iranian common bermudagrasses to find cold-tolerant accessions and evaluate their genetic and morphological variabilities. In this study, 49 accessions were collected from 18 provinces of Iran. One foreign cultivar of common bermudagrass was used as control. Morphological variation was evaluated based on 14 morphological traits to give information about taxonomic position of Iranian common bermudagrass. Data from morphological traits were evaluated to categorize all accessions as either cold sensitive or tolerant using hierarchical clustering with Ward’s method in SPSS software. Inter-Simple Sequence Repeat (ISSR) primers were employed to evaluate genetic variability of accessions. The results of our taxonomic investigation support the existence of two varieties of Cynodon dactylon in Iran: var. dactylon (hairless plant) and var. villosous (plant with hairs at leaf underside and/or upper side surfaces or exterior surfaces of sheath). All 15 primers amplified and gave clear and highly reproducible DNA fragments. In total, 152 fragments were produced, of which 144 (94.73%) being polymorphic. The polymorphic information content (PIC) values ranged from 0.700 to 0.928. The average PIC value obtained with 15 ISSR primers was 0.800, which shows that all primers were informative. Probability identity (PI) and discriminating power between all primers ranged from 0.029 to 0.185 and 0.815 to 0.971, respectively. Genetic data were converted into a binary data matrix. NTSYS software was used for data analysis. Clustering was done by the unweighted pair-group method with arithmetic averages and principle coordinate analysis, separated the accessions into six main clusters. According to both morphological and genetic diversity investigations of accessions, they can be clustered into three groups: cold sensitive, cold semi-tolerant, and cold tolerant. The most cold-tolerant accessions were: Taft, Malayear, Gorgan, Safashahr, Naein, Aligoudarz, and the foreign cultivar. This study may provide useful information for further breeding programs on common bermudagrass. Selected genotypes can be evaluated for other abiotic stresses such as drought and salinity.



Similar content being viewed by others
References
Akbari, M., & Salehi, H. (2018). Biochemical and physiological evaluations revealed high diversity within common bermudagrass (Cynodon dactylon [L.] Pers.) Iranian accessions under cold stress. Advances in Horticultural Science, 32 (in press).
Anderson, M. P., Taliaferro, C. M., Martin, D. L., & Anderson, C. S. (2001). Comparative DNA profiling of U-3 turf bermudagrass strains. Crop Science, 41, 1184–1189.
Anderson, J. A., & Taliaferro, C. M. (2002). Freeze tolerance of seed-producing turf bermudagarsses. Crop Science, 42, 190–192.
Anderson, J. A., Taliaferro, C. M., & Martin, D. L. (2003). Longer exposure durations increase freeze damage to bermudagrasses. Crop Science, 43, 973–977.
Arumuganathan, K., Tallury, S. P., Fraser, M. L., Bruneau, A., & Qu, R. (1999). Nuclear DNA content of thirteen turfgrass species by flow cytometry. Crop Science, 39, 1518–1521.
Davis, P. H. (1985). Flora of Turkey and the east Aegean islands (p. 579). Edinburgh, UK: Edinburgh University Press.
Devitt, D. A., Bowman, D. C., & Schulte, P. J. (1993). Response of Cynodon dactylon to prolonged water deficits under saline conditions. Plant and Soil, 148, 239–251.
Dipaola, J. M., & Beard, J. B. (1992). Physiological effects of temperature stress. In D. V. Waddington, R. N. Carrow, & R. C. Shearman (Eds.), Turfgrass Agronomy Monograph (Vol. 32, pp. 231–262). Madison, WI: American Society of Agronomy.
Downton, W. J. (1975). The occurrence of C4 photosynthesis among plants. Photosynthetica, 9, 96–105.
Etemadi, N., Sayed-tabatabaei, B. E., Zamani, Z., Razmjoo, K. H., Khalighi, A., & Lessani, H. (2006). Evaluation of diversity among Cynodon dactylon (L.) Pers. using RAPD markers. Journal of Agriculture and Biology, 8, 198–202.
Finger, F. L., Lannes, S. E., Schuelter, A. R., Doege, G., Comerlato, A. P., Goncalves, L. S. A., et al. (2010). Genetic diversity of Capsicum chinensis (Solanaceae) accessions based on molecular markers and morphological and agronomic traits. Genetics and Molecular Research, 9, 1852–1864.
Hajjar, R., & Hodgkin, T. (2007). The use of wild relatives in crop improvement: A survey of developments over the last 20 years. Euphytica, 156, 1–13.
Harlan, J. R., & de Wet, J. M. J. (1969). Sources of variation in Cynodon dactylon (L.) Pers. Crop Science, 9, 774–778.
Harlan, J. R. (1970). Geographic distribution of the species of Cynodon L.C. Rich (Graminae). East African Agricultural and Forestry Journal, 36, 220–226.
Hodkinson, T. R., Chase, M. W., & Renvoize, S. A. (2002). Characterization of a genetic resource collection for Miscanthus (Saccharinae, Andropogoneae, Poaceae) using AFLP and ISSR PCR. Annals of Botany, 89, 627–636.
Holm, L. G., Plunknett, D. L., Pancho, J. V., & Herberger, J. P. (1991). The world’s worst weeds: Distribution and biology (p. 609). Malabar, Florida: Krieger Publishing Company.
Huang, C. Q., Liu, G. D., Bai, C. J., Wang, W. Q., Zhou, S., & Yu, D. Q. (2010). Estimation of genetic variation in Cynodon dactylon accessions using the ISSR technique. Biochemical Systematics and Ecology, 38, 993–999.
Huang, C. Q., Liu, G., Bai, C., & Wang, W. (2014). Genetic analysis of 430 Chinese Cynodon dactylon accessions using sequence-related amplified polymorphism markers. International Journal of Molecular Sciences, 15, 19134–19146.
Ibitayo, O. O., & Butler, J. D. (1981). Cold hardiness of bermudagrass and Paspalum vaginatum SW. HortScience, 16, 683–684.
Jian, S. G., Tang, T., Zhong, Y., & Shi, S. H. (2004). Variation in inter-simple sequencerepeat (ISSR) in mangrove and non-mangrove population of Heritieralittoralis (Sterculiaceae) from China and Australia. Aquatic Botany, 79, 75–86.
Kamps, T. L., Williams, N. R., Ortega, V. M., Chamusco, K. C., Harris-Shultz, K., Scully, B. T., et al. (2011). DNA polymorphisms at bermudagrass microsatellite loci and their use in genotype fingerprinting. Crop Science, 51, 1122–1131.
Karaca, M., Saha, S., Zipf, A., Jenkins, J. N., & Lang, D. J. (2002). Genetic diversity among bermudagrass (Cynodon spp.): Evidence from chloroplast and nuclear DNA fingerprinting. Crop Science, 42, 2118–2127.
Karp, A., Edwards, K. J., Bruford, M., Funk, S., Vosman, B., Morgante, M., et al. (1997). Molecular technologies for biodiversity evaluation: Opportunities and challenges. Nature Biotechnology, 15, 625–629.
Karpinski, S., Wingsle, G., Karpinska, B., & Hallgren, J. (2002). Low-temperature stress and antioxidant defense mechanisms in higher plants. In D. Inze & M. V. Montagu (Eds.), Oxidative stress in plants (pp. 69–104). London, UK: Taylor & Francis.
Kenworthy, K. E., Taliaferro, C. M., Carver, B. F., Martin, D. L., Anderson, J. A., & Bell, G. E. (2006). Genetic variation in Cynodon transvaalensis Burtt-Davy. Crop Science, 46, 2376–2381.
Kearns, R., Zhou, Y., Fukai, S., Ye, C., Loch, D., Godwin, I., et al. (2009). Eco-turf: Water use efficient turfgrasses from Australian biodiversity. Acta Horticulturae, 829, 113–118.
Kissmann, K. (1991). Plantas infestantes e nocivas. Basf Brasileira, 2, 317–321.
Koster, K. L., & Leopold, A. C. (1988). Sugars and desiccation tolerance. Plant Physiology, 88, 829–832.
Li, H., Liu, L., Lou, Y., Hu, T., & Fu, J. (2011). Genetic diversity of Chinese natural bermudagrass (Cynodon dactylon) germplasm using ISSR markers. Scientia Horticulturae, 127, 555–561.
Luo, N., Liu, J., Yu, X., & Jiang, Y. (2011). Natural variation of drought response in Brachypodium distachyon. Physiologia Plantarum, 141, 19–29.
Majidi, M. M., & Mirlohi, M. (2010). Genetic similarities among Iranian populations of Festuca, Lolium, Bromus and Agropyron using amplified fragments length polymorphism (AFLP) markers. Iranian Journal of Biotechnology, 8, 16–23.
McCarty, L. B. (2005). Best golf course management practices. Upper Saddle River, NJ: Prentice Hall.
Mohammadi Farsani, T., Etemadi, N., Sayed-Tabatabaei, B., & Talebi, M. (2012). Assessment of genetic diversity of bermudagrass (Cynodon dactylon) using ISSR markers. International Journal of Molecular Sciences, 13, 383–392.
Mohsen, H., & Ali, F. (2008). (. Study of genetic polymorphism of Artemisia herba-alba from Tunisia using ISSR markers. African Journal of Biotechnology, 7, 44–50.
Munshaw, G. C., Ervin, E. H., Shang, C., Askew, S. D., Zhang, X., & Lemus, R. W. (2006). Influence of late-season iron, nitrogen, and seaweed extract on fall color retention and cold tolerance of four bermudagrass cultivars. Crop Science, 46, 273–283.
Nasiri, A., Saiedi, H., & Rahiminejad, M. R. (2012). Morphological variation and taxonomic conclusion of Cynodon dactylon (L.) Pers. in Iran. Asian Journal of Plant Sciences, 11, 62–69.
Oliviera, E. J., Padua, J. G., Zucchi, M. I., & Venkovsky, R. (2006). Origin, evolution and genome distribution of microsatellites. Biology, 29, 294–307.
Pollefeys, P., & Bousquet, J. (2003). Molecular genetic diversity of the French-American grapevine hybrids cultivated in North America. Genome, 46, 1037–1048.
Raina, S. N., Rani, V., Kojima, T., Ogihara, Y., Singh, K. P., & Devarumath, R. M. (2001). RAPD and ISSR fingerprints for analysis of genetic diversity, varietal identification, and phylogenetic relationships in peanut (Arachis hypogaea) cultivars and wild species. Genome, 44, 763–772.
Rohlf, F. J. (1998). NTSYSpc: Numerical taxonomy and multivariate analysis system. Version 2.02.. New York: Exeter Publications.
Roodt, R., Spies, J. J., & Burger, T. H. (2002). Preliminary DNA fingerprinting of the turfgrass Cynodon dactylon (Poaceae: Chloridoideae). Bothalia, 32, 117–122.
Rozhevits, R. Y., & Shishkin, B. K. (1934). Gramineae. In Komarov, VL. (Ed.), Flora of the U.S.S.R. Vol. 2. Koeltz Scientific Books, USA, pp. 227–228.
Salehi, M., Salehi, H., Niazi, A., & Ghobadi, C. (2014). Convergence of goals: phylogenetical, morphological, and physiological characterization of tolerance to drought stress in tall fescue (Festuca arundinacea Schreb.). Molecular Biotechnology, 56, 248–257.
Senthil Kumar, R., Parthiban, K., & Govinda Rao, M. (2009). Molecular characterization of Jatropha genetic resources through inter-simple sequence repeat (ISSR) markers. Molecular Biology Reports, 36, 1951–1956.
Shahba, M. A., Qian, Y. L., Hughes, H. G., Koski, A. J., & Christensen, D. (2003). Relationships of soluble carbohydrates and freeze tolerance in saltgrass. Crop Science, 43, 2148–2153.
Shashikumar, K., & Nus, J. L. (1993). Cultivar and winter cover effects on bermudagrass cold acclimation and crown moisture content. Crop Science, 33, 813–817.
Shyan, Y. S., Juraimi, A. S., Rafiil, M. Y., Shabanimofrad, M., Alam, M. A., & Hakim, M. A. (2014). Genetic divergence of bermudagrass (Cynodon spp.). Population using ISSR markers. Life Science Journal, 11, 425–430.
Smolik, M., Ochmian, I., & Grajkowski, J. (2010). Genetic variability of Polish and Russian accessions of cultivated blue honeysuckle (Lonicera caerulea). Genetika, 46, 1079–1085.
Taliaferro, C. M. (2000). Bermudagrass has made great strides-and its diversity has barely been tapped. How many more superb cultivars may lurk in its germplasm? Diversity, 16, 23–24.
Thomashow, M. F. (1999). Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annual Review of Plant Physiology and Plant Molecular Biology, 50, 571–599.
Williams, K., Kubelik, J. G., Livak, A. R., Rafalski, K. J., & Tingey, S. V. (1990). DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Research, 18, 6231–6235.
Wu, Y., Taliaferro, C. M., Bai, G. H., & Anderson, M. P. (2004). AFLP analysis of Cynodon dactylon (L.) Pers. Var. dactylon genetic variation. Genome, 47, 689–696.
Xie, W., Zhang, X., Cai, H., Liu, W., & Peng, Y. (2010). Genetic diversity analysis and transferability of cereal EST-SSR markers to orchardgrass (Dactylis glomerata L.). Biochemical Systematics and Ecology, 38, 740–749.
Zehdi, S., Trifi, M., Salem, A., Rhouma, A., & Marrakchi, M. (2002). Survey of inter simple sequence repeat polymorphisms in Tunisian date palms (Phoenix dactylifera L.). Journal of Genetics and Breeding, 56, 77–83.
Zhang, X., Ervin, E. H., & LaBranche, A. J. (2006). Metabolic defense responses of seeded bermudagrass during acclimation to freezing stress. Crop Science, 46, 2598–2605.
Zhao, W. G., Zhou, Z. H., Miao, X. X., Wang, S. B., Zhang, L., Yile, P., et al. (2006). Genetic relatedness among cultivated and wild mulberry (Moraceae: Morus) as revealed by inter-simple sequence repeat (ISSR) analysis in China. Canadian Journal of Plant Science, 86, 251–257.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Akbari, M., Salehi, H. & Niazi, A. Evaluation of Diversity Based on Morphological Variabilities and ISSR Molecular Markers in Iranian Cynodon dactylon (L.) Pers. Accessions to Select and Introduce Cold-Tolerant Genotypes. Mol Biotechnol 60, 259–270 (2018). https://doi.org/10.1007/s12033-018-0068-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-018-0068-5