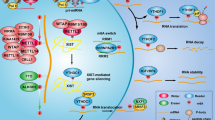
Abstract
N 6-methyladenosine (m6A) is the most abundant and reversible internal modification ubiquitously occurring in eukaryotic mRNA, albeit the significant biological roles of m6A methylation have remained largely unclear. The well-known DNA and histone methylations play crucial roles in epigenetic modification of biologic processes in eukaryotes. Analogously, the dynamic and reversible m6A RNA modification, which is installed by methyltransferase (METTL3, METTL14, and WTAP), reversed by demethylases (FTO, ALKBH5) and mediated by m6A-binding proteins (YTHDF1–3, YTHDC1), may also have a profound impact on gene expression regulation. Recent discoveries of the distributions, functions, and mechanisms of m6A modification suggest that this methylation functionally modulates the eukaryotic transcriptome to influence mRNA transcription, splicing, nuclear export, localization, translation, and stability. This reversible mRNA methylation shed light on a new dimension of post-transcriptional regulation of gene expression at the RNA level. m6A methylation also plays significant and broad roles in various physiological processes, such as development, fertility, carcinogenesis, stemness, early mortality, meiosis and circadian cycle, and links to obesity, cancer, and other human diseases. This review mainly describes the current knowledge of m6A and perspectives on future investigations.

Similar content being viewed by others
References
Machnicka, M. A., Milanowska, K., Oglou, O. O., Purta, E., Kurkowska, M., Olchowik, A., et al. (2013). MODOMICS: A database of RNA modification pathways-2013 update. Nucleic Acids Research, 41, D262–D267.
Ping, X. L., Sun, B. F., Wang, L., Xiao, W., Yang, X., Wang, W. J., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Research, 24, 177–189.
Suzuki, M. M., & Bird, A. (2008). DNA methylation landscapes: Provocative insights from epigenomics. Nature Reviews Genetics, 9, 465–476.
Jones, P. A. (2012). Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nature Reviews Genetics, 13, 484–492.
Strahl, B. D., & Allis, C. D. (2000). The language of covalent histone modifications. Nature, 403, 41–45.
Bird, A. (2001). Molecular biology—Methylation talk between histones and DNA. Science, 294, 2113–2115.
Reik, W., Dean, W., & Walter, J. (2001). Epigenetic reprogramming in mammalian development. Science, 293, 1089–1093.
Desrosie, R., Frideric, K., & Rottman, F. (1974). Identification of methylated nucleosides in messenger-RNA from Novikoff hepatoma-cells. Proceedings of the National Academy of Sciences USA, 71, 3971–3975.
Fu, Y., Dominissini, D., Rechavi, G., & He, C. (2014). Gene expression regulation mediated through reversible m(6)A RNA methylation. Nature Reviews Genetics, 15, 293–306.
Yue, Y. N., Liu, J. Z., & He, C. (2015). RNA N-6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Development, 29, 1343–1355.
Niu, Y., Zhao, X., Wu, Y. S., Li, M. M., Wang, X. J., & Yang, Y. G. (2013). N6-methyl-adenosine (m6A) in RNA: An old modification with a novel epigenetic function. Genomics, Proteomics & Bioinformatics, 11, 8–17.
Keith, G. (1995). Mobilities of modified ribonucleotides on two-dimensional cellulose thin-layer chromatography. Biochimie, 77, 142–144.
Jia, G. F., Fu, Y., Zhao, X., Dai, Q., Zheng, G. Q., Yang, Y., et al. (2011). N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nature Chemical Biology, 7, 885–887.
Zheng, G. Q., Dahl, J. A., Niu, Y. M., Fedorcsak, P., Huang, C. M., Li, C. J., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Molecular Cell, 49, 18–29.
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m(6)A RNA methylomes revealed by m(6)A-seq. Nature, 485, 201-U284.
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., & Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3′UTRs and near Stop Codons. Cell, 149, 1635–1646.
Chen, K., Lu, Z. K., Wang, X., Fu, Y., Luo, G. Z., Liu, N., et al. (2015). High-resolution N-6-methyladenosine (m(6)A) map using photo-crosslinking-assisted m(6)A sequencing. Angewandte Chemie International Edition, 54, 1587–1590.
Wei, C. M., Gershowitz, A., & Moss, B. (1975). Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell, 4, 379–386.
Perry, R. P., Kelley, D. E., Friderici, K., & Rottman, F. (1975). The methylated constituents of L cell messenger RNA: Evidence for an unusual cluster at the 5′ terminus. Cell, 4, 387–394.
Bodi, Z., Button, J. D., Grierson, D., & Fray, R. G. (2010). Yeast targets for mRNA methylation. Nucleic Acids Research, 38, 5327–5335.
Ke, S., Alemu, E. A., Mertens, C., Gantman, E. C., Fak, J. J., Mele, A., et al. (2015). A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes & Development, 29, 2037–2053.
Pan, T. (2013). N6-methyl-adenosine modification in messenger and long non-coding RNA. Trends in Biochemical Sciences, 38, 204–209.
Schwartz, S., Mumbach, M. R., Jovanovic, M., Wang, T., Maciag, K., Bushkin, G. G., et al. (2014). Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Reports, 8, 284–296.
Levis, R., & Penman, S. (1978). 5′-Terminal structures of Poly(a)+ cytoplasmic messenger-RNA and of Poly(a)+ and Poly(a)− heterogeneous nuclear-RNA of cells of dipteran Drosophila-melanogaster. Journal of Molecular Biology, 120, 487–515.
Nichols, J. L. (1979). N6-methyladenosine in maize poly(A)-containing RNA. Plant Science Letters, 15, 357–367.
Clancy, M. J., Shambaugh, M. E., Timpte, C. S., & Bokar, J. A. (2002). Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N-6-methyladenosine in mRNA: A potential mechanism for the activity of the IME4 gene. Nucleic Acids Research, 30, 4509–4518.
Krug, R. M., Morgan, M. A., & Shatkin, A. J. (1976). Influenza viral mRNA contains internal N6-methyladenosine and 5′-terminal 7-methylguanosine in cap structures. Journal of Virology, 20, 45–53.
Beemon, K., & Keith, J. (1977). Localization of N6-methyladenosine in Rous-Sarcoma virus genome. Journal of Molecular Biology, 113, 165–179.
Csepany, T., Lin, A., Baldick, C. J, Jr, & Beemon, K. (1990). Sequence specificity of mRNA N6-adenosine methyltransferase. The Journal of Biological Chemistry, 265, 20117–20122.
Batista, P. J., Molinie, B., Wang, J. K., Qu, K., Zhang, J. J., Li, L. J., et al. (2014). m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell, 15, 707–719.
Schwartz, S., Agarwala, S. D., Mumbach, M. R., Jovanovic, M., Mertins, P., Shishkin, A., et al. (2013). High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell, 155, 1409–1421.
Geula, S., Moshitch-Moshkovitz, S., Dominissini, D., Mansour, A. A., Kol, N., Salmon-Divon, M., et al. (2015). m(6)A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science, 347, 1002–1006.
Deng, X., Chen, K., Luo, G. Z., Weng, X., Ji, Q., Zhou, T., et al. (2015). Widespread occurrence of N6-methyladenosine in bacterial mRNA. Nucleic Acids Research, 43, 6557–6567.
Luo, G. Z., MacQueen, A., Zheng, G., Duan, H., Dore, L. C., Lu, Z., et al. (2014). Unique features of the m6A methylome in Arabidopsis thaliana. Nature Communications, 5, 5630.
Narayan, P., & Rottman, F. M. (1988). An invitro system for accurate methylation of internal adenosine residues in messenger-RNA. Science, 242, 1159–1162.
Bokar, J. A., Shambaugh, M. E., Polayes, D., Matera, A. G., & Rottman, F. M. (1997). Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N-6-adenosine)-methyltransferase. RNA, 3, 1233–1247.
Bokar, J. A. (2005). The biosynthesis and functional roles of methylated nucleosides in eukaryotic mRNA. In H. Grosjean (Ed.), Fine-tuning of RNA functions by modification and editing (pp. 141–177). Berlin: Springer.
Zhong, S., Li, H., Bodi, Z., Button, J., Vespa, L., Herzog, M., et al. (2008). MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. The Plant Cell, 20, 1278–1288.
Hongay, C. F., & Orr-Weaver, T. L. (2011). Drosophila inducer of MEiosis 4 (IME4) is required for Notch signaling during oogenesis. Proceedings of the National Academy of Sciences U S A, 108, 14855–14860.
Bujnicki, J. M., Feder, M., Radlinska, M., & Blumenthal, R. M. (2002). Structure prediction and phylogenetic analysis of a functionally diverse family of proteins homologous to the MT-A70 subunit of the human mRNA:m(6)A methyltransferase. Journal of Molecular Evolution, 55, 431–444.
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nature Chemical Biology, 10, 93–95.
Wang, Y., Li, Y., Toth, J. I., Petroski, M. D., Zhang, Z., & Zhao, J. C. (2014). N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nature Cell Biology, 16, 191–198.
Little, N. A., Hastie, N. D., & Davies, R. C. (2000). Identification of WTAP, a novel Wilms’ tumour 1-associating protein. Human Molecular Genetics, 9, 2231–2239.
Horiuchi, K., Umetani, M., Minami, T., Okayama, H., Takada, S., Yamamoto, M., et al. (2006). Wilms’ tumor 1-associating protein regulates G(2)/M transition through stabilization of cyclin A2 mRNA. Proceedings of the National Academy of Sciences USA, 103, 17278–17283.
Horiuchi, K., Kawamura, T., Iwanari, H., Ohashi, R., Naito, M., Kodama, T., et al. (2013). Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. Journal of Biological Chemistry, 288, 33292–33302.
Agarwala, S. D., Blitzblau, H. G., Hochwagen, A., & Fink, G. R. (2012). RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLoS Genetics, 8, e1002732.
Peters, T., Ausmeier, K., & Ruther, U. (1999). Cloning of Fatso (Fto), a novel gene deleted by the Fused toes (Ft) mouse mutation. Mammalian Genome, 10, 983–986.
Fischer, J., Koch, L., Emmerling, C., Vierkotten, J., Peters, T., Bruning, J. C., et al. (2009). Inactivation of the Fto gene protects from obesity. Nature, 458, 894-U810.
Church, C., Moir, L., McMurray, F., Girard, C., Banks, G. T., Teboul, L., et al. (2010).Overexpression of Fto leads to increased food intake and results in obesity. Nature Genetics, 42, 1086-U1147.
Frayling, T. M., Timpson, N. J., Weedon, M. N., Zeggini, E., Freathy, R. M., Lindgren, C. M., et al. (2007). A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science, 316, 889–894.
Dina, C., Meyre, D., Gallina, S., Durand, E., Korner, A., Jacobson, P., et al. (2007). Variation in FTO contributes to childhood obesity and severe adult obesity. Nature Genetics, 39, 724–726.
Scuteri, A., Sanna, S., Chen, W. M., Uda, M., Albai, G., Strait, J., et al. (2007). Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genetics, 3, 1200–1210.
Gerken, T., Girard, C. A., Tung, Y. C. L., Webby, C. J., Saudek, V., Hewitson, K. S., et al. (2007). The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science, 318, 1469–1472.
Boissel, S., Reish, O., Proulx, K., Kawagoe-Takaki, H., Sedgwick, B., Yeo, G. S. H., et al. (2009). Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations. American Journal of Human Genetics, 85, 106–111.
Wang, X., Zhu, L., Chen, J., & Wang, Y. (2015). mRNA m(6)A methylation downregulates adipogenesis in porcine adipocytes. Biochemical and Biophysical Research Communications, 459, 201–207.
Kurowski, M. A., Bhagwat, A. S., Papaj, G., & Bujnicki, J. M. (2003). Phylogenomic identification of five new human homologs of the DNA repair enzyme AlkB. BMC Genomics, 4, 48.
Falnes, P. O., Johansen, R. F., & Seeberg, E. (2002). AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature, 419, 178–182.
Trewick, S. C., Henshaw, T. F., Hausinger, R. P., Lindahl, T., & Sedgwick, B. (2002). Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature, 419, 174–178.
Fu, Y., Jia, G., Pang, X., Wang, R. N., Wang, X., Li, C. J., et al. (2013). FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nature Communications, 4, 1798.
Tahiliani, M., Koh, K. P., Shen, Y. H., Pastor, W. A., Bandukwala, H., Brudno, Y., et al. (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science, 324, 930–935.
Ito, S., D’Alessio, A. C., Taranova, O. V., Hong, K., Sowers, L. C. & Zhang, Y. (2010). Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature, 466, 1129-U1151.
Ito, S., Shen, L., Dai, Q., Wu, S. C., Collins, L. B., Swenberg, J. A., et al. (2011). Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science, 333, 1300–1303.
Zhao, X., Yang, Y., Sun, B. F., Shi, Y., Yang, X., Xiao, W., et al. (2014). FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Research, 24, 1403–1419.
Gulati, P., Cheung, M. K., Antrobus, R., Church, C. D., Harding, H. P., Tung, Y. C. L., et al. (2013). Role for the obesity-related FTO gene in the cellular sensing of amino acids. Proceedings of the National Academy of Sciences USA, 110, 2557–2562.
Vujovic, P., Stamenkovic, S., Jasnic, N., Lakic, I., Djurasevic, S. F., Cvijic, G., et al. (2013). Fasting induced cytoplasmic Fto expression in some neurons of rat hypothalamus. PLoS One, 8, e63694.
Zheng, G. Q., Dahl, J. A., Niu, Y. M., Fu, Y., Klungland, A., Yang, Y. G., et al. (2013). Sprouts of RNA epigenetics: The discovery of mammalian RNA demethylases. RNA Biology, 10, 915–918.
Xu, C., Liu, K., Tempel, W., Demetriades, M., Aik, W., Schofield, C. J., et al. (2014). Structures of human ALKBH5 demethylase reveal a unique binding mode for specific single-stranded N-6-methyladenosine RNA demethylation. Journal of Biological Chemistry, 289, 17299–17311.
Aik, W., Scotti, J. S., Choi, H., Gong, L. Z., Demetriades, M., Schofield, C. J., et al. (2014). Structure of human RNA N-6-methyladenine demethylase ALKBH5 provides insights into its mechanisms of nucleic acid recognition and demethylation. Nucleic Acids Research, 42, 4741–4754.
Wang, X., Lu, Z. K., Gomez, A., Hon, G. C., Yue, Y. N., Han, D. L., et al. (2014). N-6-methyladenosine-dependent regulation of messenger RNA stability. Nature, 505, 117–120.
Stoilov, P., Rafalska, I., & Stamm, S. (2002). YTH: A new domain in nuclear proteins. Trends in Biochemical Sciences, 27, 495–497.
Zhang, Z. Y., Theler, D., Kaminska, K. H., Hiller, M., de la Grange, P., Pudimat, R., et al. (2010). The YTH domain is a novel RNA binding domain. Journal of Biological Chemistry, 285, 14701–14710.
Sheth, U., & Parker, R. (2003). Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science, 300, 805–808.
Cardelli, M., Marchegiani, F., Cavallone, L., Olivieri, F., Giovagnetti, S., Mugianesi, E., et al. (2006). A polymorphism of the YTHDF2 gene (1p35) located in an Alu-rich genomic domain is associated with human longevity. The Journal of Gerontology A, 61, 547–556.
Xu, C., Wang, X., Liu, K., Roundtree, I. A., Tempel, W., Li, Y. J., et al. (2014). Structural basis for selective binding of m(6)A RNA by the YTHDC1 YTH domain. Nature Chemical Biology, 10, 927–929.
Wang, X., Zhao, B. S., Roundtree, I. A., Lu, Z. K., Han, D. L., Ma, H. H., et al. (2015). N-6-methyladenosine modulates messenger RNA translation efficiency. Cell, 161, 1388–1399.
Abdelmohsen, K., & Gorospe, M. (2010). Posttranscriptional regulation of cancer traits by HuR. Wires RNA, 1, 214–229.
Filippova, N., Yang, X. H., King, P., & Nabors, L. B. Z. (2012). Phosphoregulation of the RNA-binding protein Hu antigen R (HuR) by Cdk5 Affects centrosome function. Journal of Biological Chemistry, 287, 32277–32287.
Wan, Y., Kertesz, M., Spitale, R. C., Segal, E., & Chang, H. Y. (2011). Understanding the transcriptome through RNA structure. Nature Reviews Genetics, 12, 641–655.
Dreyfuss, G., Kim, V. N., & Kataoka, N. (2002). Messenger-RNA-binding proteins and the messages they carry. Nature Reviews Molecular Cell Biology, 3, 195–205.
Ray, D., Kazan, H., Cook, K. B., Weirauch, M. T., Najafabadi, H. S., Li, X., et al. (2013). A compendium of RNA-binding motifs for decoding gene regulation. Nature, 499, 172–177.
Liu, N., Dai, Q., Zheng, G. Q., He, C., Parisien, M., & Pan, T. (2015). N-6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature, 518, 560–564.
Konig, J., Zarnack, K., Rot, G., Curk, T., Kayikci, M., Zupan, B., et al. (2010). iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nature Structural & Molecular Biology, 17, 909-U166.
McCloskey, A., Taniguchi, I., Shinmyozu, K., & Ohno, M. (2012). hnRNP C tetramer measures RNA length to classify RNA polymerase II transcripts for export. Science, 335, 1643–1646.
Liu, N., Parisien, M., Dai, Q., Zheng, G. Q., He, C., & Pan, T. (2013). Probing N-6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA, 19, 1848–1856.
Theler, D., & Allain, F. H. T. (2015). Molecular biology RNA modification does a regulatory two-step. Nature, 518, 492–493.
Thomson, J. A., Itskovitz-Eldor, J., Shapiro, S. S., Waknitz, M. A., Swiergiel, J. J., Marshall, V. S., et al. (1998). Embryonic stem cell lines derived from human blastocysts. Science, 282, 1145–1147.
Dunn, S. J., Martello, G., Yordanov, B., Emmott, S., & Smith, A. G. (2014). Defining an essential transcription factor program for naive pluripotency. Science, 344, 1156–1160.
Young, R. A. (2011). Control of the embryonic stem cell state. Cell, 144, 940–954.
Chen, T., Hao, Y. J., Zhang, Y., Li, M. M., Wang, M., Han, W., et al. (2015). m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell, 16, 289–301.
Fustin, J. M., Doi, M., Yamaguchi, Y., Hida, H., Nishimura, S., Yoshida, M., et al. (2013). RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell, 155, 793–806.
Akhtar, R. A., Reddy, A. B., Maywood, E. S., Clayton, J. D., King, V. M., Smith, A. G., et al. (2002). Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Current Biology, 12, 540–550.
Koike, N., Yoo, S. H., Huang, H. C., Kumar, V., Lee, C., Kim, T. K., et al. (2012). Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science, 338, 349–354.
Jackson, R. J., Hellen, C. U. T., & Pestova, T. V. (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nature Reviews Molecular Cell Biology, 11, 113–127.
Hinnebusch, A. G. (2014). The scanning mechanism of eukaryotic translation initiation. Annual Review of Biochemistry, 83, 779–812.
Zhou, J., Wan, J., Gao, X. W., Zhang, X. Q., Jaffrey, S. R. and Qian, S. B. (2015). Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature, 526, 591-U332.
Meyer, K. D., Patil, D. P., Zhou, J., Zinoviev, A., Skabkin, M. A., Elemento, O., et al. (2015). 5’ UTR m(6)A promotes cap-independent translation. Cell, 163, 999–1010.
Sibbritt, T., Patel, H. R., & Preiss, T. (2013). Mapping and significance of the mRNA methylome. Wires RNA, 4, 397–422.
Loos, R. J. F., & Bouchard, C. (2008). FTO: The first gene contributing to common forms of human obesity. Obesity Reviews, 9, 246–250.
Loos, R. J. F., & Yeo, G. S. H. (2014). The bigger picture of FTO-the first GWAS-identified obesity gene. Nature Reviews Endocrinology, 10, 51–61.
Alarcon, C. R., Lee, H., Goodarzi, H., Halberg, N., & Tavazoie, S. F. (2015). N-6-methyladenosine marks primary microRNAs for processing. Nature, 519, 482–485.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 31372320) and the Special Fund for Cultivation and Breeding of New Transgenic Organism (No. 2014ZX0800949B).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wu, R., Jiang, D., Wang, Y. et al. N 6-Methyladenosine (m6A) Methylation in mRNA with A Dynamic and Reversible Epigenetic Modification. Mol Biotechnol 58, 450–459 (2016). https://doi.org/10.1007/s12033-016-9947-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-016-9947-9