Abstract
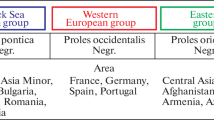
Grapevine is the most economically important and widely cultivated fruit crop in the world. Molecular markers have been used on Vitis vinifera to distinguish among both varieties and clones. Microsatellites are used to fingerprint varieties and several other techniques, reported in many papers, are used to analyze the differences among clones, but it is not available in the literature as a well defined strategy to screen a large number of Vitis cultivars. In fact, it is often necessary to use different techniques to investigate the genetic variability in different grapevine varieties and a proposed technique is used to study a cultivar, which is often not suitable for either the study of another cultivar or compare the genetic relationship among various cultivars. We describe here a strategy used for the analysis of several grapevine cultivars to describe a universal method to obtain DNA polymorphisms of Vitis vinifera genotypes from the same cultivar by using amplified fragment length polymorphism (AFLP), selective amplification of microsatellite polymorphic loci (SAMPL), microsatellites AFLP (M-AFLP), and ISSR molecular markers. The strategy here adopted permitted both to identify different biotypes (i.e., Primitivo), accessions (i.e., Garnacha tinta), and clones (i.e., Callet, Manto Negro, Moll) among the variability of same variety and to correlate the genetic differences to their geographical origins (i.e., Garnacha tinta; Malvasia nera di Brindisi/Lecce) or morphological traits (i.e., Malvasia of Candia). Here is also described the application of the protocol that allows to highlight the genetic variability accumulated during centuries of cultivations and selections of the same variety in different environments by vine growers.









Similar content being viewed by others
References
Mullis, L. G., Bouquet, A., & Williams, L. E. (1992). The biology of the grapevine. Cambridge, MA: Cambridge University Press.
Meneghetti, S., Costacurta, A., Morreale, G., & Calò, A. (2011). Study of intravarietal genetic variability in grapevine cultivars by PCR-derived molecular markers and correlations with the geographic origin. Molecular Biotechnology. doi:10.1007/s12033-011-9403-9.
Galet, P. (2000). Dictionnaire encyclopédique des cépages. Paris: Hachette.
Meneghetti, S., Calò, A., Costacurta, A., Frare, E., & Crespan, M. (2010). Valutazione della variabilità intra-varietale in vite ai fini dell’identificazione clonale—Evaluation of the intra-varietal variability for the clones identification. Rivista Viticoltura Enologia, 1-2-3-4, 93–103.
De Crescenzi, P. (1805). Trattato dell’agricoltura (B. de Rossi, Trans). Milano: Società Tipografica dei Classici Italiani.
Odart, G. (1874). Traité de cépages. Paris: Librairie agricole.
Meneghetti, S., Poljuha, D., Frare, E., Costacurta A., Morreale, G., Bavaresco, L., & Calò, A. (2011). Inter- and Intra-varietal genetic variability in Malvasias. Molecular Biotechnology. doi:10.1007/s12033-011-9423-5.
de Martínez Toda, F., & Sancha, J. C. (1997). Ampelographical characterization of red Vitis vinifera L. cultivars preserved in Rioja. Bull de l’OIV, 70, 220–234.
Bachmann, O., & Blaich, R. (1988). Isoelectric focusing of grapevine peroxidases as a tool for ampelography. Vitis, 27, 147–155.
Calò, A., & Costacurta, A. (2004). Dei vitigni italici. Treviso: Ed. Matteo.
Costacurta, A., Egger, E., Storchi, P., Crespan, M., Dilani, N., Sensi, E., & Carraro, R. (2001). Caratterizzazione molecolare, ampelografica ed ampelometrica di 30 accessioni di Vitis vinifera L. riferibili al Sangiovese. International Conference of Sangiovese, Firenze (Italy) 8–10th March 2000, pp. 113–120. Press Effeemme Lito.
Costacurta, A., Calò, A., Carraro, R., Lorenzoni, C., & Giust, M. (2000). VII International Symposium on grapevine genetics and breeding, Montpellier, France. ISHS Acta Horticulturae 528.
Martí, C., Casanova, J., Montaner, C., & Badia, D. (2006). Ampelometric study of mature leaves from two indigenous Vitis cultivars grown in Somontano de Barbastro. Journal of Wine Research, 17(3), 185–194.
Zapata, C., Deléens, E., Chaillou, S., & Magné, C. (2004). Mobilisation and distribution of starch and total N in two grapevine cultivars differing in their susceptibility to shedding. Functional Plant Biology, 31(11), 1127–1135.
Chervin, C., Tira-Umphon, A., Chatelet, P., Jauneau, A., Boss, P. K., & Tesniere, C. (2009). Ethylene and other stimuli affect expression of the UDP glucose-flavonoid 3-O-glucosyltransferase in a non-climacteric fruit. Vitis, 48, 11–16.
D’Onofrio, C., Cox, A., Davies, C., & Boss, P. K. (2009). Induction of secondary metabolism in grape cell cultures by jasmonates. Functional Plant Biology, 36, 323–338.
Kalua, C. M., & Boss, P. K. (2009). Evolution of volatile compounds during the development of Cabernet Sauvignon grapes (Vitis vinifera L.). Journal of Agricultural and Food Chemistry, 57, 3818–3830.
Godevac, D., Teševiæ, V., Velièkoviæ, M., Vujisiæ, L., Vajs, V., & Milosavljeviæ, S. (2010). Polyphenolic compounds in seeds from some grape cultivars grown in Serbia. Journal of the Serbian Chemical Society, 75(12), 1641–1652.
Keyzers, R. A., & Boss, P. K. (2010). Changes in the volatile compound production of fermentations made from musts with increasing grape content. Journal of Agricultural and Food Chemistry, 58, 1153–1164.
Wolf, W. H. (1976). Identification of grape varieties by isozyme banding patterns. American Journal of Enology and Viticulture, 27(2), 68–73.
Schwennesen, J., Mielke, E. A., & Wolfe, W. H. (1982). Identification of seedless table grape cultivars and a bud sport with berry isozymes. Hortscience, 17(3), 366–368.
Loukas, M., Stavrakakis, M. N., & Krimbas, C. B. (1983). Inheritance of polymorphic isoenzymes in grape cultivars. The Journal of Heredity, 74(3), 181–183.
Altube, H., Cabello, F., & Ortiz, J. M. (1991). Caracterización de variedades y portainjertos de vid mediante isoenzimas de los sarmientos. Vitis, 30, 203–212.
Silvestroni, O., & Intrieri, C. (1995). Ampelometric assessment of clonal variability in the Sangiovese winegrape cultivar. International Symposium on Clonal Selection, 20–21 June, Portland, OR, USA (pp. 137–142). American Society for Enology and Viticulture.
Calò, A., Costacurta, A., Cancellier, S., & Forti, R. (1990). Garnacha, Grenache, Cannonao, Tocai rosso, un unico vitigno. Vignevini, 9, 45–48.
Royo, J. B., Cabello, F., Miranda, S., Gogorcena, Y., González, J., Moreno, S., et al. (1997). The use of isoenzymes in characterization of grapevines (Vitis vinifera L.). Influence of the environment and time of sampling. Scientia Horticulturae, 69, 145–155.
Tessier, C., David, J., This, P., Boursiquot, J. M., & Charrier, A. (1999). Optimization of the choice of molecular markers for varietal identification in Vitis vinifera L. Theoretical and Applied Genetics, 89, 171–177.
Pelsy, F., Hocquigny, S., Moncada, X., Barbeau, G., Forget, D., Hinrichsen, P., et al. (2010). An extensive study of the genetic diversity within seven French wine grape variety collections. Theoretical and Applied Genetics, 120(6), 1219–1231.
Sefc, K. M., Regner, F., Turetschek, E., Glössl, J., & Steinkellner, H. (1999). Identification of microsatellite sequences in Vitis riparia and their applicability for genotyping of different Vitis species. Genome, 42, 367–373.
Meneghetti, S., Costacurta, A., Crespan, M., Maul, E., Hack, R., & Regner, F. (2009). Deepening inside the homonyms of Wildbacher by means of SSR markers. Vitis, 48(3), 123–129.
Lefort, F., & Roubelakis-Angelakis, K. A. (2001). Genetic comparison of Greek cultivars of Vitis vinifera L. by nuclear microsatellite profiling. American Journal of Enology and Viticulture, 52(2), 101–108.
Cipriani, G., Spadotto, A., Jurman, I., Di Gaspero, G., Crespan, M., Meneghetti, S., et al. (2010). The SSR-based profile of 1005 grapevine accessions uncovers new synonymy and parentages and reveals a large admixture among varieties of different geographic origin. Theoretical and Applied Genetics, 121(8), 1569–1585.
Jallion, O., Aury, J. M., Noel, B., Policriti, A., Clepet, C., Casagrande, A., et al. (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature, 449, 463–468.
Cipriani, G., Marrazzo, M. T., Di Gaspero, G., Pfeiffer, A., Morgante, M., & Testolin, R. (2008). A set of microsatellite markers with long core repeat optimized for grape (Vitis spp.) genotyping. BMC Plant Biology, 8, 127.
Meneghetti, S., Costacurta, A., & Calò, A. (2010). Le développement de méthodes moléculaires pour l’identification des cultivars dans la vigne. OIV Oral communication N. CI-GENET 2010-03-07. Organisation Internationale de la Vigne et du Vin—Section Experts en Génétique, Paris, le 17 mars 2010.
Cretazzo, E., Meneghetti, S., De Andrés, M. T., Frare, E., Gaforio, L., & Cifre, J. (2010). Clone differentiation and varietal identification by means of SSR, AFLP, SAMPL and M-AFLP in order to assist the clonal selection of grapevine. The case of study of Manto Negro, Callet and Moll, autochthonous cultivar of Majorca. Annals of Applied Biology, 157(2), 213–227.
Meneghetti, S., Costacurta, A., Frare, E., Da Rold, G., Migliaro, D., Morreale, G., et al. (2011). Clones identification and genetic characterization of Garnacha grapevine by means of different PCR-derived marker systems. Molecular Biotechnology, 48(3), 244–254.
Moreno, S., Gogorcena, Y., & Ortiz, J. M. (1995). The use of RAPD markers for identification of cultivated grapevine (Vitis vinifera L.). Scientia Horticulturae, 62(4), 237–243.
Böhm, A., & Zyprian, E. (1998). RAPD marker in grapevine (Vitis spp.) similar to plant retrotransposons. Plant Cell Reports, 17(5), 415–421.
Regner, F., Wiedeck, E., & Stadlbauer, A. (2000). Differentiation and identification of White Riesling clones by genetic markers. Vitis, 39(3), 103–107.
Herrera, R., Cares, V., Wilkinson, M. J., & Calidari, P. D. S. (2002). Characterisation of genetic variation between Vitis vinifera cultivars from central Chile using RAPD and Inter Simple Sequence Repeat markers. Euphytica, 124(1), 139–145.
Karataş, H., & Ağaoğlu, Y. S. (2010). RAPD analysis of selected local Turkish grape cultivars (Vitis vinifera). Genetics and Molecular Research, 9(4), 1980–1986.
Moreno, S., Martin, J., & Ortiz, J. (1998). Inter simple sequence repeats PCR for characterization of closely related grapevine germplasm. Euphytica, 101, 117–125.
Tamhankar, S. A., Argade, N. C., More, M. N., Dhanorkar, V. M., Patil, S. G., Rao, V. S., et al. (2008). DNA profiling of the grape varieties grown in India using ISSR markers. Acta Horticulturae, 785, 147–152.
Owens, C. L. (2003). SNP detection and genotyping in Vitis. Acta Horticulturae, 603, 139–140.
Troggio, M., Malacarne, G., Coppola, G., Segala, C., Cartwright, D. A., Pindo, M., et al. (2007). A dense Single Nucleotide Polymorphism based genetic linkage map of grapevine (Vitis vinifera L.) anchoring Pinot noir bacterial artificial chromosome Contigs. Genetics, 176, 2637–2650.
Troggio, M., Malacarne, G., Vezzulli, S., Faes, G., Salmaso, M., & Velasco, R. (2008). Comparison of different methods for SNP detection in grapevine. Vitis, 47(1), 21–30.
Labra, M., Imazio, S., Grassi, F., Rossoni, M., & Sala, F. (2004). Vine-1 retrotransposon-based sequence-specific amplified polymorphism for Vitis vinifera L. genotyping. Plant Breeding, 123(2), 180–185.
Pelsey, F., Schehrer, L., & Merdinoglu, D. (2002). Development of grapevine molecular markers based on retrotransposons. Acta Horticulturae, 603, 83–87.
D’Onofrio, C., De Lorenzis, G., Giordani, T., Natali, L., Scalabrelli, G., & Cavallini, A. (2009). Retrotransposon-based molecular markers in grapevine species and cultivars identification and phylogenetic analysis. Acta Horticulturae, 827, 45–52.
Imazio, S., Labra, M., Grassi, F., Winfield, M., Bardini, M., & Scienza, A. (2002). Molecular tools (SSR, AFLP, MSAP) for clone identification: The case of the grapevine cultivar ‘Traminer’. Plant Breeding, 121(6), 531–535.
Arroyo-García, R., Ruiz-García, L., Bolling, L., Ocete, R., López, M. A., Arnold, C., et al. (2006). Multiple origins of cultivated grapevine (Vitis vinifera L. ssp. sativa) based on chloroplast DNA polymorphisms. Molecular Ecology, 15(12), 3707–3714.
Hunt, H. V., Lawes, M. C., Bower, M. A., Haeger, J. W., & Howe, C. J. (2010). A banned variety was the mother of several major wine grapes. Biology Letters, 6(3), 367–369.
Wegscheider, E., Benjak, A., & Forneck, A. (2009). Clonal variation in Pinot noir revealed by S-SAP involving universal retrotransposon-based sequences. American Journal of Enology and Viticulture, 60(1), 104–109.
Değirmenci Karataþ, D., Kunter, B., Coppola, G., & Velasco, R. (2010). Analysis of polymorphism based on SSCP markers in gamma-irradiated (Co60) grape (Vitis vinifera) varieties. Genetics and Molecular Research, 9(4), 2357–2363.
Ulanovskya, S., Gogorcenab, Y., Martínez de Todac, F., & Ortiz, J. M. (2002). Use of molecular markers in detection of synonymies and homonymies in grapevines (Vitis vinifera L.). Scientia Horticulturae, 92, 241–254.
Meneghetti, S., Costacurta, A., & Calò, A. (2009). Evaluation of the intra-varietal variability for the clones identification (II). OIV Oral communication N. CI-GENET 03.2009-07.1. Organisation Internationale de la Vigne et du Vin—Section Experts en Génétique, Paris, le 18 mars 2009.
Coletta, A., Crespan, M., Costacurta, A., Caputo, A. R., Taurisano, C., Meneghetti, S., et al. (2006). Preliminary investigations on Malvasia nera di Lecce and Malvasia nera di Brindisi varieties. Rivista Viticoltura Enologia, 2(3), 51–56.
Calò, A., Masi, G., Tarricone, L., Costacurta, A., Meneghetti, S., Crespan, M., et al. (2008). Search for Primitivo (V. vinifera L.) variability in Apulia. Rivista Viticoltura Enologia, 1, 3–13.
Sefc, M. K., Lopes, M. S., Lefort, F., Botta, R., Roubelakis-Angelakis, K. A., Ibáñez, J., et al. (2000). Microsatellite variability in grapevine cultivars from different European regions and evaluation of assignment testing to assess the geographic origin of cultivars. Theoretical and Applied Genetics, 100, 498–505.
Costacurta, A., Calò, A., Giannetto, S., Giust, M., Comandini, F., & Meneghetti S. (2009) Experience of typologies identification of Malvasia bianca di Candia using morphological traits and molecular markers. Oral communication at the III Malvasias International Symposium, La Palma, Canary Islands, March 27–28th 2009.
Cervera, M. T., Cabezas, J. A., Sancha, J. C., de Martínez Toda, F., & Martínez-Zapater, J. M. (1998). Application of AFLPs to the characterization of grapevine Vitis vinifera L. genetic resources. A case of study with accessions from Rioja. Theoretical and Applied Genetics, 97, 51–59.
Fanizza, G., Chaabane, R., Ricciardi, L., & Resta, P. (2003). Analysis of a spontaneous mutant and selected clones of cv. Italia (Vitis vinifera) by AFLP markers. Vitis, 42(1), 27–30.
Blaich, R., Konradi, J., Rühl, E., & Forneck, A. (2007). Assessing genetic variation among Pinot noir (Vitis vinifera L.) clones with AFLP markers. American Journal of Enology and Viticulture, 58(4), 526–529.
López, M., Cid, N., González, M. V., Cuenca, B., Prado, M. J., & Rey, M. (2009). Microsatellite and AFLP analysis of autochthonous grapevine cultivars from Galicia (Spain). American Journal of Enology and Viticulture, 60(2), 215–222.
Wolf, T., Cabezas, J. A., & Martínez-Zapater, J. M. (2003). Genetic characterization of closely related rootstocks varieties based on AFLP and SAMPL markers. Acta Horticulturae, 603, 291–300.
Application Notes of Genomics, Life Science Robotics: Automated genomic DNA extraction from grapevine, AN_NS8Plant_HamMicrolabStar.pdf. http://www.mnnet.com/Portals/8/attachments/Redakteure_Bio/ApplicationNotes/AN_NS8Plant_HamMicrolabStar.pdf.
Barcaccia, G., Meneghetti, S., Albertini, E., Triest, L., & Lucchin, M. (2003). Linkage mapping in tetraploid willows: Segregation of molecular markers and estimation of linkage phases support an allotetraploid structure for Salix alba × Salix fragilis interspecific hybrids. Heredity, 90, 169–180.
Meneghetti, S., Barcaccia, G., Paiero, P., & Lucchin, M. (2007). Genetic characterization of Salix alba L. and Salix fragilis L. by means of different PCR-derived marker systems. Plant Biosystems, 141(3), 283–291.
Albertini, E., Porceddu, A., Marconi, G., Barcaccia, G., Pallottini, L., & Falcinelli, M. (2003). Microsatellite-AFLP for genetic mapping of complex polyploids. Genome, 46, 824–832.
Dice, L. R. (1945). Measures of the amount of ecological association between species. Ecology, 26, 297–302.
Sneath, P. H. A., & Sokal, R. R. (1973). Numerical taxonomy (p. 513). San Francisco, CA: Freeman.
Powell, W., Machray, G. C., & Provan, J. (1996). Polymorphism revealed by simple sequence repeats. Trends in Plant Science, 1, 215–222.
Rohlf, F. J., & Sokal, R. R. (1981). Comparing numerical taxonomic studies. Systematic Zoology, 30, 459–490.
Tuimala, J. (2006). A primer to phylogenetic analysis using the PHYLIP package (5th ed., p. 55). The Author and CSC. Espoo, Finland: Scientific Computing Ltd. http://www.ku.edu.np/biotech/bioinfodata/phylip2.pdf. Accessed 3 Aug 2010.
Maletić, E., Pejić, I., Karoglan Kontić, J., Piljac, J., Dangl, G. S., Vokurta, A., et al. (2004). Zinfandel, Dobričić, and Plavac mali: The genetic relationship among three cultivars of the Dalmatian coast of Croatia. American Journal of Enology and Viticulture, 55, 174–180.
Jung, A. (2007). Zinfandel, the story continues: New insights to its ancient variety history from a German point of view. Rivista Viticoltura Enologia, 3, 37–58.
Calò, A., Costacurta, A., Maraš, V., Meneghetti, S., & Crespan, M. (2008). Molecular correlation of Zinfandel (Primitivo) with Austrian, Croatian, and Hungarian cultivars and Kratošija, an additional synonym. American Journal of Enology and Viticulture, 59(2), 205–209.
Antonacci, D. (2006). Grapevines of Apulia. Bari: Mario Adda Editor.
Crespan, M., Coletta, A., Crupi, P., Giannetto, S., & Antonacci, D. (2008). Malvasia nera di Brindisi/Lecce’ grapevine cultivar (Vitis vinifera L.) originated from ‘Negroamaro’ and ‘Malvasia bianca lunga’. Vitis, 47(4), 205–212.
Acknowledgments
This study is supported by both “ASER” and “IDENTIVIT” research grants from Ministero delle Politiche Agricole Alimentari e Forestali (MiPAAF), Rome, Italy. We are very grateful to Dr Giacomo Morreale (researcher at the CRA-VIT) for their indispensable cooperation and support. We would also like to thank Dr. Angelo Costacurta (former Director of CRA-VIT) and Dr Enrica Frare (CRA-VIT) for their comments and suggestions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Meneghetti, S., Calò, A. & Bavaresco, L. A Strategy to Investigate the Intravarietal Genetic Variability in Vitis vinifera L. for Clones and Biotypes Identification and to Correlate Molecular Profiles with Morphological Traits or Geographic Origins. Mol Biotechnol 52, 68–81 (2012). https://doi.org/10.1007/s12033-011-9475-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-011-9475-6