Abstract
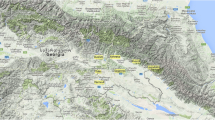
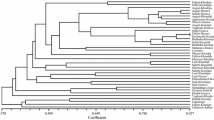
The genetic grapevine intravarietal variability will be analyzed by PCR-derived marker systems. In particular, the object of the investigation will be the clonal variations of Malvasia nera di Brindisi/Lecce, Negroamaro and Primitivo, also known as Zinfandel, which are three grapevine varieties cultivated in Apulia region (Italy). In order to assess varietal identity of the samples, 132 DNA tests were performed by amplifying 16 SSR loci. The study of the intravarietal variability was performed using AFLPs, SAMPLs, ISSRs, and M-AFLPs. The application of the above-mentioned techniques allowed both to discriminate all genotypes of the three cultivars and to distinguish the accessions of each cultivar sampled from different geographic cultivation areas. Furthermore, the study of biotypes cultivated in different geographical environments of Salento (i.e., Apulia region) allowed important correlations between molecular marker variability and phenotypic traits. These results are suggesting both to focus our attention on the effects of the environment on the genotype and to consider, as a practical consequence, the importance of preserving autochthon grapevine biotypes found in different areas to truly preserve the richness of the germplasm. Thus, more accurate DNA studies give new information that can be extremely useful to the vine nurseries for the correct choice (i.e., supported by more accurate intravarietal variability analysis) of the grape multiplication materials.







Similar content being viewed by others
References
Arrigo, N., & Arnold, C. (2007). Naturalised Vitis Rootstocks in Europe and consequences to Native Wild Grapevine. PLoS One, 2–6, 521.
Clarke, O. (2001). Encyclopedia of grape (pp. 91–100). Orlando: Harcourt Books.
Columella. (1781). De re rustica (book III, 9, 1). (J. M. Gesner, Trans.). L. Junii Moderati Columellae edition.
De Crescenzi, P. (1805). Trattato dell’agricoltura. (Bastiano de Rossi, Trans.). Società tipografica dei Classici Italiani. Milan: Italy.
Odart, G. (1854). Ampélographie universelle ou traité de cépages les plus estimès (III ed.). Paris: Librairie agricole.
Galet, P. (1979). A practical ampelography: Grapevine identification. Ithaca, New York: University Press.
Calò, A., Costacurta, A., Cancellier, S., & Forti, R. (1990). Garnacha, Grenache, Cannonao, Tocai rosso, un unico vitigno. Vignevini, 9, 45–48.
Calò, A., & Costacurta, A. (2004). Dei vitigni italici. Treviso: Ed. Matteo.
de Martínez Toda, F., & Sancha, J. C. (1997). Ampelographical characterization of red Vitis vinifera L. cultivars preserved in Rioja. Bull de l’OIV, 70, 220–234.
Loukas, M., Stavrakakis, M. N., & Krimbas, C. B. (1983). Inheritance of polymorphic isoenzymes in grape cultivars. The Journal of Heredity, 74–3, 181–183.
Altube, H., Cabello, F., & Ortiz, J. M. (1991). Caracterización de variedades y portainjertos de vid mediante isoenzimas de los sarmientos. Vitis, 30, 203–212.
Bachmann, O., & Blaich, R. (1988). Isoelectric focusing of grapevine peroxidases as a tool for ampelography. Vitis, 27, 147–155.
Wolf, W. H. (1976). Identification of grape varieties by isozyme banding patterns. American Journal of Enology and Viticulture, 27–2, 68–73.
Schwennesen, J., Mielke, E. A., & Wolfe, W. H. (1982). Identification of seedless table grape cultivars and a bud sport with berry isozymes. Hortscience, 17–3, 366–368.
Royo, J. B., Cabello, F., Miranda, S., Gogorcena, Y., González, J., Moreno, S., et al. (1997). The use of isoenzymes in characterization of grapevines (Vitis vinifera L.) Influence of the environment and time of sampling. Scientia Horticulturae, 69, 145–155.
Tessier, C., David, J., This, P., Boursiquot, J. M., & Charrier, A. (1999). Optimization of the choice of molecular markers for varietal identification in Vitis vinifera L. Theoretical and Applied Genetics, 89, 171–177.
Pelsy, F., Hocquigny, S., Moncada, X., Barbeau, G., Forget, D., Hinrichsen, P., et al. (2010). An extensive study of the genetic diversity within seven French wine grape variety collections. Theoretical and Applied Genetics, 120–6, 1219–1231.
Techera, G., Jubany, A., de Ponce León, S., Boido, I., Dellacassa, E., Carrau, E., et al. (2004). Molecular diversity (SSR) within clones of cv. Tannat (Vitis vinifera). Vitis, 43–44, 179–185.
Galet, P. (2000). Dictionnaire encyclopédique des cépages. Paris: Hachette.
Meneghetti, S., Costacurta, A., Crespan, M., Maul, E., Hack, R., & Regner, F. (2009). Deepening inside the homonyms of Wildbacher by means of SSR markers. Vitis, 48–3, 123–129.
Stavrakakis, M., & Loukas, M. (1983). The between and within grape cultivars genetic variation. Scientia Horticulturae, 19, 321–334.
Bachmann, K. (1994). Molecular markers in plant ecology. New Phytologist, 126, 403–418.
Chaparro, J. X., Goldy, R. G., Mowrey, B. D., & Werner, D. J. (1989). Identification of Vitis vinifera × Muscardinia rotundifolia small hybrids by starch gel electrophoresis. Hortscience, 24, 128–130.
Moreno, S., Gogorcena, Y., & Ortiz, J. M. (1995). The use of RAPD markers for identification of cultivated grapevine (Vitis vinifera L.). Scientia Horticulturae, 62–4, 237–243.
Böhm, A., & Zyprian, E. (1998). RAPD marker in grapevine (Vitis spp.) similar to plant retrotransposons. Plant Cell Reports, 17–5, 415–421.
Imazio, S., Labra, M., Grassi, F., Winfield, M., Bardini, M., & Scienza, A. (2002). Molecular tools (SSR, AFLP, MSAP) for clone identification: The case of the grapevine cultivar ‘Traminer’. Plant Breeding, 121–6, 531–535.
Pelsey, F., Schehrer, L., & Merdinoglu, D. (2002). Development of grapevine molecular markers based on retrotransposons. Acta Horticulturae, 603, 83–87.
Owens, C. L. (2003). SNP detection and genotyping in Vitis. Acta Horticulturae, 603, 139–140.
Labra, M., Imazio, S., Grassi, F., Rossoni, M., & Sala, F. (2004). Vine-1 retrotransposon-based sequence-specific amplified polymorphism for Vitis vinifera L. genotyping. Plant Breeding, 123–2, 180–185.
Meneghetti, S., Costacurta A., Frare, E., Da Rold, G., Migliaro, D., Morreale, G., Crespan, M., Sotés, V., Calò, A. (2011). Clones Identification and genetic characterization of Garnacha grapevine by means of different pcr-derived marker systems. Molecular Biotechnology. doi:10.1007/s12033-010-9365-3.
D’ Onofrio, C., De Lorenzis, G., Giordani, T., Natali, L., Scalabrelli, G., & Cavallini, A. (2009). Retrotransposon-based molecular markers in grapevine species and cultivars identification and phylogenetic analysis. Acta Horticulturae, 827, 45–52.
Cervera, M. T., Cabezas, J. A., Sancha, J. C., de Martínez Toda, F., & Martínez-Zapater, J. M. (1998). Application of AFLPs to the characterization of grapevine Vitis vinifera L. genetic resources. A case of study with accessions from Rioja. Theoretical and Applied Genetics, 97–1(2), 51–59.
Fanizza, G., Chaabane, R., Ricciardi, L., & Resta, P. (2003). Analysis of a spontaneous mutant and selected clones of cv. Italia (Vitis vinifera) by AFLP markers. Vitis, 42, 27–30.
Blaich, R., Konradi, J., Rühl, E., & Forneck, A. (2007). Assessing genetic variation among Pinot noir (Vitis vinifera L.) clones with AFLP markers. American Journal of Enology and Viticulture, 58–4, 526–529.
Cretazzo, E., Meneghetti, S., De Andrés, M. T., Frare, E., Gaforio, L., & Cifre, J. (2010). Clone differentiation and varietal identification by means of SSR, AFLP, SAMPL and M-AFLP in order to assist the clonal selection of grapevine. The case of study of Manto Negro, Callet and Moll, autochthonous cultivar of Majorca. Annals of Applied Biology, 157–2, 213–227.
Regner, F., Wiedeck, E., & Stadlbauer, A. (2000). Differentiation and identification of White Riesling clones by genetic markers. Vitis, 39–3, 103–107.
Barcaccia, G., Meneghetti, S., Albertini, E., Triest, L., & Lucchin, M. (2003). Linkage mapping in tetraploid willows: Segregation of molecular markers and estimation of linkage phases support an allotetraploid structure for Salix alba × Salix fragilis interspecific hybrids. Heredity, 90, 169–180.
Wolf, T., Cabezas, J. A., & Martínez-Zapater, J. M. (2003). Genetic characterization of closely related rootstocks varieties based on AFLP and SAMPL markers. Acta Horticulturae, 603, 291–300.
Albertini, E., Porceddu, A., Marconi, G., Barcaccia, G., Pallottini, L., & Falcinelli, M. (2003). Microsatellite-AFLP for genetic mapping of complex polyploids. Genome, 46, 824–832.
Meneghetti, S., Costacurta, A., & Calò, A. (2009). Evaluation of the intra-varietal variability for the clones identification (II). OIV Oral communication N. CI-GENET 03.2009-07.1. Organisation internationale de la vigne et du vin—Section Experts en Génétique, Paris, le 18 Mars 2009.
Sefc, M. K., Lopes, M. S., Lefort, F., Botta, R., Roubelakis-Angelakis, K. A., Ibáñez, J., et al. (2000). Microsatellite variability in grapevine cultivars from different European regions and evaluation of assignment testing to assess the geographic origin of cultivars. Theoretical and Applied Genetics, 100, 498–505.
Antonacci, D. (2006). Grapevines of Apulia. Bari: Mario Adda Editor.
Meneghetti, S., Costacurta, A., Calò, A., Sotés, V., Giannetto, S., Crespan, M. (2006). Investigation on Italian, Spanish and French Garnacha tinta genetic variability—a preliminary study. Oral communication at the XXIX OIV 2006 International Symposium, Logroño, 25-30 Junio, España.
López, M., Cid, N., González, M. V., Cuenca, B., Prado, M. J., & Rey, M. (2009). Microsatellite and AFLP Analysis of Autochthonous Grapevine Cultivars from Galicia (Spain). American Journal of Enology and Viticulture, 60–2, 215–222.
Crespan, M., Cabello, F., Giannetto, S., Ibáñez, J., Kontić, J. K., Maletić, E., et al. (2006). Malvasia delle Lipari, Malvasia di Sardegna, Greco di Gerace, Malvasia de Sitges and Malvasia dubrovačka—synonyms of an old and famous grape cultivar. Vitis, 45, 69–73.
Lacombe, T., Boursiquot, J. M., Laucou, V., Dechesne, F., Varès, D., & This, P. (2007). Relationships and genetic diversity within the accessions related to Malvasia held in the Domaine de Vassal Grape Germplasm Repository. American Journal of Enology and Viticulture, 58, 124–131.
Crespan, M., Coletta, A., Crupi, P., Giannetto, S., & Antonacci, D. (2008). Malvasia nera di Brindisi/Lecce’ grapevine cultivar (Vitis vinifera L.) originated from ‘Negroamaro’ and ‘Malvasia bianca lunga. Vitis, 47–4, 205–212.
Coletta, A., Crespan, M., Costacurta, A., Caputo, A. R., Taurisano, C., Meneghetti, S., et al. (2006). Preliminary investigations on Malvasia nera di Lecce and Malvasia nera di Brindisi varieties. Rivista Viticoltura Enologia, 2–3, 51–56.
Calò, A., Costacurta, A., Catalano, V., & Di Stefano, R. (2000). Negro Amaro precoce. Rivista Viticoltura Enologia, 3, 27–44.
Calò, A., Costacurta, A., Maraš, V., Meneghetti, S., & Crespan, M. (2008). Molecular Correlation of Zinfandel (Primitivo) with Austrian, Croatian, and Hungarian Cultivars and Kratošija, an Additional Synonym. American Journal of Enology and Viticulture, 59–2, 205–209.
Calò, A., Masi, G., Tarricone, L., Costacurta, A., Meneghetti, S., Crespan, M., et al. (2008). Search for Primitivo (V. vinifera L.) variability in Apulia. Rivista Viticoltura Enologia, 1, 3–13.
Maletić, E., Pejić, I., Karoglan Kontić, J., Piljac, J., Dangl, G. S., Vokurta, A., et al. (2004). Zinfandel, Dobričić, and Plavac mali: The genetic relationship among three cultivars of the Dalmatian coast of Croatia. American Journal of Enology and Viticulture, 55, 174–180.
Jung, A. (2007). Zinfandel, the story continues: New insights to its ancient variety history from a German point of view. Rivista Viticoltura Enologia, 3, 37–58.
Crespan, M. (2004). Evidence on the evolution of polymorphism of microsatellite markers in varieties of Vitis vinifera L. Theoretical and Applied Genetics, 108, 231–237.
Crespan, M. (2003). The parentage of Muscat of Hamburg. Vitis, 42(4), 193–197.
Thomas, M. R., & Scott, N. S. (1993). Microsatellite repeats in grapevine reveal DNA polymorphisms when analysed as sequence tagged sites (STSs). Theoretical and Applied Genetics, 86, 985–990.
Bowers, J. E., Dangl, G. S., & Meredith, C. P. (1999). Development and characterization of additional microsatellite DNA markers for grape. American Journal of Enology and Viticulture, 53, 125–130.
Barcaccia, G., Mazzuccato, A., Albertini, E., Zethof, J., Gerats, A., Pezzotti, M., et al. (1998). Inheritance of parthenogenesis in Poa pratensis L.: Auxin test and AFLP linkage analyses support monogenic control. Theoretical and Applied Genetics, 96, 74–82.
Meneghetti, S., Barcaccia, G., Paiero, P., & Lucchin, M. (2007). Genetic characterization of Salix alba L. and Salix fragilis L. by means of different PCR-derived marker systems. Plant Biosystems, 141–3, 283–291.
Van Eijk, M., De Ruiter, M., Broekhof, J., & Peleman, J. (2001). Discovery and detection of polymorphic microsatellites by microsatellite-AFLP. In Plant and animal genome IX conference (p. 143).
Dangl, G. S., Mendum, M. L., Prins, B. H., Walzer, M. A., Meredith, C. P., & Simon, C. J. (2001). Simple sequence repeat analysis of a clon-ally propagated species: A tool for managing a grape germplasm collection. Genome, 44, 432–438.
Rohlf, F. J. (2000). Numerical taxonomy and multivariate analysis system. Version 2.1. Stony Brook, NY: State University of New York.
Dice, L. R. (1945). Measures of the amount of ecological association between species. Ecology, 26, 297–302.
Sneath, P. H. A., & Sokal, R. R. (1973). Numerical Taxonomy (p. 513). San Francisco: Freeman.
Rohlf, F. J., & Sokal, R. R. (1981). Comparing numerical taxonomic studies. Systematic Zoology, 30, 459–490.
Tuimala, J. (2006). A Primer to Phylogenetic Analysis Using the PHYLIP Package (5th edn.) (p. 55). The Author and CSC Espoo: Scientific Computing Ltd. http://www.ku.edu.np/biotech/bioinfodata/phylip2.pdf.
Powell, W., Machray, G. C., & Provan, J. (1996). Polymorphism revealed by simple sequence repeats. Trends in Plant Science, 1, 215–222.
Acknowledgments
This study was part of the “ASER” project funded by Ministero delle Politiche Agricole Alimentari e Forestali (MiPAAF), Rome, Italy.
Author information
Authors and Affiliations
Corresponding author
Additional information
An erratum to this article can be found at http://dx.doi.org/10.1007/s12033-011-9410-x
Rights and permissions
About this article
Cite this article
Meneghetti, S., Costacurta, A., Morreale, G. et al. Study of Intra-Varietal Genetic Variability in Grapevine Cultivars by PCR-Derived Molecular Markers and Correlations with the Geographic Origins. Mol Biotechnol 50, 72–85 (2012). https://doi.org/10.1007/s12033-011-9403-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-011-9403-9