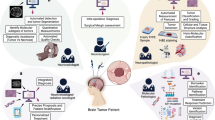
Abstract
Gliomas are one of the most devastating primary brain tumors which impose significant management challenges to the clinicians. The aggressive behaviour of gliomas is mainly attributed to their rapid proliferation, unravelled genomics and the blood–brain barrier which protects the tumor cells from chemotherapeutic regimens. Suspects of brain tumors are usually assessed by magnetic resonance imaging and computed tomography. These images allow surgeons to decide on the tumor grading, intra-operative pathology, feasibility of surgery, and treatment planning. All these data are compiled manually by physicians, wherein it takes time for the validation of results and concluding the treatment modality. In this context, the arrival of artificial intelligence in this era of personalized medicine, has proven promising performance in the diagnosis and management of gliomas. Starting from grading prediction till outcome evaluation, artificial intelligence-based forefronts have revolutionized oncological research. Interestingly, this approach has also been able to precisely differentiate tumor lesion from healthy tissues. However, till date, their utility in neuro-oncological field remains limited due to the issues pertaining to their reliability and transparency. Hence, to shed novel insights on the “clinical utility of this novel approach on glioma management” and to reveal “the black-boxes that have to be solved for fruitful application of artificial intelligence in neuro-oncology research”, we provide in this review, a succinct description of the potential gear of artificial intelligence-based avenues in glioma treatment and the barriers that impede their rapid implementation in neuro-oncology.



Similar content being viewed by others
References
Hanif F, Muzaffar K, Perveen K, Malhi SM, Simjee SU. Glioblastoma multiforme: a review of its epidemiology and pathogenesis through clinical presentation and treatment. Asian Pac J Cancer Prev. 2017;18:3–9.
Vigneswaran K, Neill S, Hadjipanayis CG. Beyond the World Health Organization grading of infiltrating gliomas: advances in the molecular genetics of glioma classification. Ann Transl Med. 2015;3:95.
Harder BG, Blomquist MR, Wang J, Kim AJ, Woodworth GF, Winkles JA, et al. Developments in blood-brain barrier penetrance and drug repurposing for improved treatment of glioblastoma. Front Oncol. 2018;8:462.
Davis ME. Glioblastoma: overview of disease and treatment. Clin J Oncol Nurs. 2016;20:S2-8.
Sanghvi D. Post-treatment imaging of high-grade gliomas. Indian J Radiol Imaging. 2015;25:102–8.
Hamilton JG, Genoff Garzon M, Westerman JS, Shuk E, Hay JL, Walters C, et al. “A Tool, Not a Crutch”: patient perspectives about IBM Watson for Oncology trained by Memorial Sloan Kettering. JOP. 2019;15:e277–88.
Choi YI, Chung J, Kim KO, Kwon KA, Kim YJ, Park DK, et al. Concordance rate between clinicians and Watson for oncology among patients with advanced gastric cancer: early, real-world experience in Korea. Can J Gastroenterol Hepatol. 2019;2019:8072928.
Lee W-S, Ahn SM, Chung J-W, Kim KO, Kwon KA, Kim Y, et al. Assessing concordance with Watson for oncology, a cognitive computing decision support system for colon cancer treatment in Korea. JCO Clin Cancer Inform. 2018. https://doi.org/10.1200/CCI.17.00109.
Shahid N, Rappon T, Berta W. Applications of artificial neural networks in health care organizational decision-making: a scoping review. PLoS ONE. 2019;14:e0212356.
Choy G, Khalilzadeh O, Michalski M, Do S, Samir AE, Pianykh OS, et al. Current applications and future impact of machine learning in radiology. Radiology. 2018;288:318–28.
Yamashita R, Nishio M, Do RKG, Togashi K. Convolutional neural networks: an overview and application in radiology. Insights Imaging. 2018;9:611–29.
Noel Sharkey. Alan turing: the experiment that shaped artificial intelligence. BBC News [Internet]. 2012 Jun 20 [cited 2020 Nov 14]; Available from: https://www.bbc.com/news/technology-18475646.
Reddy S. Use of artificial intelligence in healthcare delivery. eHealth—making health care smarter [Internet]. IntechOpen; 2018 [cited 2020 Nov 14]; Available from: https://www.intechopen.com/books/ehealth-making-health-care-smarter/use-of-artificial-intelligence-in-healthcare-delivery.
Sikchi SS, Sikchi S, Ali MS. Artificial intelligence in medical diagnosis. Int J Appl Eng Res. 2012;7:1539–43.
Harrer S, Shah P, Antony B, Hu J. Artificial intelligence for clinical trial design. Trends Pharmacol Sci. 2019;40:577–91.
Shimizu H, Nakayama KI. Artificial intelligence in oncology. Cancer Sci. 2020;111:1452–60.
Wirtz BW, Weyerer JC. Artificial intelligence in the public sector. In: Farazmand A, editor. Global encyclopedia of public administration, public policy, and governance [Internet]. Cham: Springer International Publishing; 2019 [cited 2020 Nov 14]. p. 1–7. Available from: https://doi.org/10.1007/978-3-319-31816-5_3701-1.
Kersting K. Machine learning and artificial intelligence: two fellow travelers on the quest for intelligent behavior in machines. Front Big Data. 2018;1:6.
Deloitte Insights State of AI in the enterprise. [Internet]. Deloitte, 2018; 2018. Available from: https://www2.deloitte.com/content/dam/insights/us/articles/4780_State-of-AI-in-the-enterprise/AICognitiveSurvey2018_Infographic.pdf.
McBee MP, Awan OA, Colucci AT, Ghobadi CW, Kadom N, Kansagra AP, et al. Deep learning in radiology. Acad Radiol. 2018;25:1472–80.
Science O-OD. Transforming skewed data for machine learning [Internet]. Medium. 2019 [cited 2020 Nov 14]. Available from: https://medium.com/@ODSC/transforming-skewed-data-for-machine-learning-90e6cc364b0.
Carrasco OC. Support vector machines for classification [Internet]. Medium. 2019 [cited 2020 Nov 14]. Available from: https://towardsdatascience.com/support-vector-machines-for-classification-fc7c1565e3.
Lan H. Decision trees and random forests for classification and regression pt.1 [Internet]. Medium. 2020 [cited 2020 Nov 14]. Available from: https://towardsdatascience.com/decision-trees-and-random-forests-for-classification-and-regression-pt-1-dbb65a458df.
Yiu T. Understanding random forest [Internet]. Medium. 2019 [cited 2020 Nov 14]. Available from: https://towardsdatascience.com/understanding-random-forest-58381e0602d2.
Lee E-J, Kim Y-H, Kim N, Kang D-W. Deep into the brain: artificial intelligence in stroke imaging. J Stroke. 2017;19:277–85.
Ghorbanzadeh O, Blaschke T, Gholamnia K, Meena SR, Tiede D, Aryal J. Evaluation of different machine learning methods and deep-learning convolutional neural networks for landslide detection. Remote Sens. 2019;11:196.
Fakoor R, Ladhak F, Nazi A, Huber M. Using deep learning to enhance cancer diagnosis and classification. In: Proceedings of the ICML Workshop on the Role of Machine Learning in Transforming Healthcare. 2013.
Azer SA. Deep learning with convolutional neural networks for identification of liver masses and hepatocellular carcinoma: a systematic review. World J Gastrointest Oncol. 2019;11:1218–30.
Brownlee J. A gentle introduction to transfer learning for deep learning [Internet]. Machine Learning Mastery. 2017 [cited 2020 Nov 14]. Available from: https://machinelearningmastery.com/transfer-learning-for-deep-learning/.
Berthold E. AI and the robotics revolution [Internet]. Curious. 2020 [cited 2020 Nov 14]. Available from: https://www.science.org.au/curious/technology-future/ai-and-robotics-revolution.
IFR. Global industrial robot sales doubled over the past five years [Internet]. IFR International Federation of Robotics. [cited 2020 Nov 14]. Available from: https://ifr.org/ifr-press-releases/news/global-industrial-robot-sales-doubled-over-the-past-five-years.
Kumar Y. Artificial intelligence & robotics—synthetic brain in action (2018 Sep 14). Available from SSRN: https://ssrn.com/abstract=3325115.
Davenport TH, Glaser J. Just-in-time delivery comes to knowledge management. Harv Bus Rev. 2002;80(107–11):126.
Zanagnolo V, Garbi A, Achilarre MT, Minig L. Robot-assisted surgery in gynecologic cancers. J Minim Invasive Gynecol. 2017;24:379–96.
Kalantari F, Rajaeih S, Daneshvar A, Karbasi Z, Mahdi Salem M. Robotic surgery of head and neck cancers, a narrative review. Eur J Transl Myol. 2020;30:8727.
Zorn KC, Gofrit ON, Steinberg GD, Shalhav AL. Evolution of robotic surgery in the treatment of localized prostate cancer. Curr Treat Options Oncol. 2007;8:197–210.
Obuchowski NA, Bullen JA. Statistical considerations for testing an AI algorithm used for prescreening lung CT images. Contemp Clin Trials Commun. 2019;16:100434.
Hajian-Tilaki K. Receiver operating characteristic (ROC) curve analysis for medical diagnostic test evaluation. Caspian J Intern Med. 2013;4:627–35.
Wang Z, Wang E, Zhu Y. Image segmentation evaluation: a survey of methods. Artif Intell Rev. 2020;53:5637–74.
Jonathan Bush. How AI is taking the scut work out of health care. Harvard Business Review [Internet]. 2018 Mar 5 [cited 2020 Nov 14]; Available from: https://hbr.org/2018/03/how-ai-is-taking-the-scut-work-out-of-health-care.
Zhou N, Zhang C, Lv H, Hao C, Li T, Zhu J, et al. Concordance study between IBM Watson for oncology and clinical practice for patients with cancer in China. Oncologist. 2019;24:812–9.
Eliza Strickland. How IBM Watson overpromised and underdelivered on AI Health Care—IEEE Spectrum [Internet]. IEEE Spectrum: Technology, Engineering, and Science News. 2019 [cited 2020 Nov 14]. Available from: https://spectrum.ieee.org/biomedical/diagnostics/how-ibm-watson-overpromised-and-underdelivered-on-ai-health-care.
Rysavy M. Evidence-based medicine: a science of uncertainty and an art of probability. AMA J Ethics. 2013;15:4–8.
Roger Montti. Free Google AI image analysis tool [Internet]. Search Engine Journal. 2019 [cited 2020 Nov 14]. Available from: https://www.searchenginejournal.com/google-cloud-vision-tool/304237/.
Sanksshep Mahendra. Artificial intelligence in healthcare [Internet]. Artificial Intelligence, IoT, and Robotics. 2020 [cited 2020 Nov 14]. Available from: https://www.aiplusinfo.com/blog/ai-in-healthcare.
Fisher R, Pusztai L, Swanton C. Cancer heterogeneity: implications for targeted therapeutics. Br J Cancer. 2013;108:479–85.
Barbara Rosen. Foundation medicine and flatiron health collaborate to develop first-in-class data platform to accelerate precision medicine for cancer | Flatiron Health [Internet]. 2014 [cited 2020 Nov 14]. Available from: https://flatiron.com/press/press-release/foundation-medicine-and-flatiron-health-collaborate-to-develop-first-in-class-data-platform-to-accelerate-precision-medicine-for-cancer/.
Rudie JD, Rauschecker AM, Bryan RN, Davatzikos C, Mohan S. Emerging applications of artificial intelligence in neuro-oncology. Radiology. 2019;290:607–18.
Brandes AA. State-of-the-art treatment of high-grade brain tumors. Semin Oncol. 2003;30:4–9.
Alieva M, van Rheenen J, Broekman MLD. Potential impact of invasive surgical procedures on primary tumor growth and metastasis. Clin Exp Metastasis. 2018;35:319–31.
Kong K, Rowlands CJ, Varma S, Perkins W, Leach IH, Koloydenko AA, et al. Diagnosis of tumors during tissue-conserving surgery with integrated autofluorescence and Raman scattering microscopy. Proc Natl Acad Sci USA. 2013;110:15189–94.
NCI Staff. Artificial intelligence expedites brain tumor diagnosis—National Cancer Institute [Internet]. 2020 [cited 2020 Nov 14]. Available from: https://www.cancer.gov/news-events/cancer-currents-blog/2020/artificial-intelligence-brain-tumor-diagnosis-surgery.
Francis Collins. Artificial intelligence speeds brain tumor diagnosis [Internet]. NIH Director’s Blog. 2020 [cited 2020 Nov 14]. Available from: https://directorsblog.nih.gov/2020/01/14/artificial-intelligence-speeds-brain-tumor-diagnosis/.
Wu C-C, Guo W-Y, Chen M-H, Ho DMT, Hung ASC, Chung H-W. Direct measurement of the signal intensity of diffusion-weighted magnetic resonance imaging for preoperative grading and treatment guidance for brain gliomas. J Chin Med Assoc. 2012;75:581–8.
Li-Chun Hsieh K, Chen C-Y, Lo C-M. Quantitative glioma grading using transformed gray-scale invariant textures of MRI. Comput Biol Med. 2017;83:9283480.
Wen PY, Huse JT. 2016 World Health Organization classification of central nervous system tumors. CONTINUUM. 2017;23:1531.
Tian Q, Yan L-F, Zhang X, Zhang X, Hu Y-C, Han Y, et al. Radiomics strategy for glioma grading using texture features from multiparametric MRI. J Magn Reson Imaging. 2018;48:1518–28.
Mao Y, Liao W, Cao D, Zhao L, Wu X, Kong L, et al. An artificial neural network model for glioma grading using image information. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2018;43:1315–22.
Ranjith G, Parvathy R, Vikas V, Chandrasekharan K, Nair S. Machine learning methods for the classification of gliomas: initial results using features extracted from MR spectroscopy. Neuroradiol J. 2015;28:106–11.
Zhang X, Yan L-F, Hu Y-C, Li G, Yang Y, Han Y, et al. Optimizing a machine learning based glioma grading system using multi-parametric MRI histogram and texture features. Oncotarget. 2017;8:47816–30.
Brat D, Prayson R, Ryken T, Olson J. Diagnosis of malignant glioma: role of neuropathology. J Neurooncol. 2008;89:287–311.
Sonoda Y, Shibahara I, Kawaguchi T, Saito R, Kanamori M, Watanabe M, et al. Association between molecular alterations and tumor location and MRI characteristics in anaplastic gliomas. Brain Tumor Pathol. 2015;32:99–104.
Kristensen BW, Priesterbach-Ackley LP, Petersen JK, Wesseling P. Molecular pathology of tumors of the central nervous system. Ann Oncol. 2019;30:1265–78.
Modrek AS, Golub D, Khan T, Bready D, Prado J, Bowman C, et al. Low-grade astrocytoma mutations in IDH1, P53 and ATRX cooperate to block differentiation of human neural stem cells via repression of SOX2. Cell Rep. 2017;21:1267–80.
Wu S, Meng J, Yu Q, Li P, Fu S. Radiomics-based machine learning methods for isocitrate dehydrogenase genotype prediction of diffuse gliomas. J Cancer Res Clin Oncol. 2019;145:543–50.
Zhang B, Chang K, Ramkissoon S, Tanguturi S, Bi WL, Reardon DA, et al. Multimodal MRI features predict isocitrate dehydrogenase genotype in high-grade gliomas. Neuro Oncol. 2017;19:109–17.
Korfiatis P, Kline TL, Lachance DH, Parney IF, Buckner JC, Erickson BJ. Residual deep convolutional neural network predicts MGMT methylation status. J Digit Imaging. 2017;30:622–8.
Han L, Kamdar MR. MRI to MGMT: predicting methylation status in glioblastoma patients using convolutional recurrent neural networks. Pac Symp Biocompt. 2018; 23: 331–42. Available from: https://www.worldscientific.com/doi/abs/https://doi.org/10.1142/9789813235533_0031.
Zhou M, Scott J, Chaudhury B, Hall L, Goldgof D, Yeom KW, et al. Radiomics in brain tumor: image assessment, quantitative feature descriptors, and machine-learning approaches. Am J Neuroradiol. 2018;39:208–16.
Scheie D, Andresen PA, Cvancarova M, Bø AS, Helseth E, Skullerud K, et al. Fluorescence in situ hybridization (FISH) on touch preparations: a reliable method for detecting loss of heterozygosity at 1p and 19q in oligodendroglial tumors. Am J Surg Pathol. 2006;30:828–37.
Parsons DW, Jones S, Zhang X, Lin JC–H, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12.
Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–73.
Jansen NL, Schwartz C, Graute V, Eigenbrod S, Lutz J, Egensperger R, et al. Prediction of oligodendroglial histology and LOH 1p/19q using dynamic [18F] FET-PET imaging in intracranial WHO grade II and III gliomas. Neuro Oncol. 2012;14:1473–80.
Hartmann C, Hentschel B, Wick W, Capper D, Felsberg J, Simon M, et al. Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: implications for classification of gliomas. Acta Neuropathol. 2010;120:707–18.
Fellah S, Caudal D, Paula AMD, Dory-Lautrec P, Figarella-Branger D, Chinot O, et al. Multimodal MR imaging (diffusion, perfusion, and spectroscopy): is it possible to distinguish oligodendroglial tumor grade and 1p/19q codeletion in the pretherapeutic diagnosis? Am J Neuroradiol. 2013;34:1326–33.
Bourdillon P, Hlaihel C, Guyotat J, Guillotton L, Honnorat J, Ducray F, et al. Prediction of anaplastic transformation in low-grade oligodendrogliomas based on magnetic resonance spectroscopy and 1p/19q codeletion status. J Neurooncol. 2015;122:529–37.
Iwadate Y, Shinozaki N, Matsutani T, Uchino Y, Saeki N. Molecular imaging of 1p/19q deletion in oligodendroglial tumours with 11C-methionine positron emission tomography. J Neurol Neurosurg Psychiatry. 2016;87:1016–21.
Hu N, Richards R, Jensen R. Role of chromosomal 1p/19q co-deletion on the prognosis of oligodendrogliomas: a systematic review and meta-analysis. Interdiscip Neurosurg. 2016;5:58–63.
Akkus Z, Ali I, Sedlář J, Agrawal JP, Parney IF, Giannini C, et al. Predicting deletion of chromosomal arms 1p/19q in low-grade gliomas from MR images using machine intelligence. J Digit Imaging. 2017;30:469–76.
Chang P, Grinband J, Weinberg BD, Bardis M, Khy M, Cadena G, et al. Deep-learning convolutional neural networks accurately classify genetic mutations in gliomas. Am J Neuroradiol. 2018;39:1201–7.
Liu J, Chen F, Pan C, Zhu M, Zhang X, Zhang L, et al. A cascaded deep convolutional neural network for joint segmentation and genotype prediction of brainstem gliomas. IEEE Trans Biomed Eng. 2018;65:1943–52.
Bilgin C, Demir C, Nagi C, Yener B. Cell-graph mining for breast tissue modeling and classification. In: 2007 29th Annual International Conference of the IEEE Engineering in Medicine and Biology Society. 2007. p. 5311–4.
Sertel O, Kong J, Catalyurek UV, Lozanski G, Saltz JH, Gurcan MN. Histopathological image analysis using model-based intermediate representations and color texture: follicular lymphoma grading. J Sign Process Syst Sign Image Video Technol. 2008;55:169.
Gurcan MN, Boucheron LE, Can A, Madabhushi A, Rajpoot NM, Yener B. Histopathological image analysis: a review. IEEE Rev Biomed Eng. 2009;2:147–71.
Marchevsky AM, Wick MR. Evidence-based medicine, medical decision analysis, and pathology. Hum Pathol. 2004;35:1179–88.
Rodenacker K, Bengtsson E. A feature set for cytometry on digitized microscopic images. Anal Cell Pathol. 2003;25:1–36.
Bera K, Schalper KA, Rimm DL, Velcheti V, Madabhushi A. Artificial intelligence in digital pathology—new tools for diagnosis and precision oncology. Nat Rev Clin Oncol. 2019;16:703–15.
Abas FS, Gokozan HN, Goksel B, Otero JJ, Gurcan MN. Intraoperative neuropathology of glioma recurrence: cell detection and classification. Medical Imaging 2016: Digital Pathology. International Society for Optics and Photonics; 2016. p. 979109. Available from: https://www.spiedigitallibrary.org/conference-proceedings-of-spie/9791/979109/Intraoperative-neuropathology-of-glioma-recurrence-cell-detection-and-classification/10.1117/12.2216448.short.
Fukuma K, Prasath VBS, Kawanaka H, Aronow BJ, Takase H. A study on nuclei segmentation, feature extraction and disease stage classification for human brain histopathological images. Proc Comput Sci. 2016;96:1202–10.
Wang X, Wang D, Yao Z, Xin B, Wang B, Lan C, et al. Machine learning models for multiparametric glioma grading with quantitative result interpretations. Front Neurosci. 2019;11:1046.
Yonekura A, Kawanaka H, Prasath VBS, Aronow BJ, Takase H. Automatic disease stage classification of glioblastoma multiforme histopathological images using deep convolutional neural network. Biomed Eng Lett. 2018;8:321–7.
Ker J, Bai Y, Lee HY, Rao J, Wang L. Automated brain histology classification using machine learning. J Clin Neurosci. 2019;66:239–45.
Gordillo N, Montseny E, Sobrevilla P. State of the art survey on MRI brain tumor segmentation. Magn Reson Imaging. 2013;31:1426–38.
LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–44.
Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88.
Lotan E, Jain R, Razavian N, Fatterpekar GM, Lui YW. State of the art: machine learning applications in glioma imaging. Am J Roentgenol. 2018;212:26–37.
Tustison NJ, Shrinidhi KL, Wintermark M, Durst CR, Kandel BM, Gee JC, et al. Optimal symmetric multimodal templates and concatenated random forests for supervised brain tumor segmentation (simplified) with ANTsR. Neuroinformatics. 2015;13:209–25.
Pereira S, Pinto A, Alves V, Silva CA. Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging. 2016;35:1240–51.
Havaei M, Davy A, Warde-Farley D, Biard A, Courville A, Bengio Y, et al. Brain tumor segmentation with deep neural networks. Med Image Anal. 2017;35:18–31.
Kamnitsas K, Ledig C, Newcombe VFJ, Simpson JP, Kane AD, Menon DK, et al. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal. 2017;36:61–78.
Cui S, Mao L, Jiang J, Liu C, Xiong S. Automatic semantic segmentation of brain gliomas from MRI images using a deep cascaded neural network. J Healthc Eng. 2018;2018:4940593.
Blanc-Durand P, Gucht AVD, Schaefer N, Itti E, Prior JO. Automatic lesion detection and segmentation of 18F-FET PET in gliomas: a full 3D U-Net convolutional neural network study. PLoS ONE. 2018;13:e0195798.
Li Z, Wang Y, Yu J, Shi Z, Guo Y, Chen L, et al. Low-grade glioma segmentation based on CNN with fully connected CRF. J Healthc Eng. 2017;2017:9283480.
Hambardzumyan D, Bergers G. Glioblastoma: defining tumor niches. Trends Cancer. 2015;1:252–65.
Sengupta A, Agarwal S, Gupta PK, Ahlawat S, Patir R, Gupta RK, et al. On differentiation between vasogenic edema and non-enhancing tumor in high-grade glioma patients using a support vector machine classifier based upon pre and post-surgery MRI images. Eur J Radiol. 2018;106:199–208.
Bonte S, Goethals I, Van Holen R. Machine learning based brain tumour segmentation on limited data using local texture and abnormality. Comput Biol Med. 2018;98:39–47.
Garzon-Muvdi T, Kut C, Li X, Chaichana KL. Intraoperative imaging techniques for glioma surgery. Future Oncol. 2017;13:1731–45.
Fabelo H, Halicek M, Ortega S, Shahedi M, Szolna A, Piñeiro JF, et al. Deep learning-based framework for in vivo identification of glioblastoma tumor using hyperspectral images of human brain. Sensors. 2019;19:920.
Zhang H, Wang R, Yu Y, Liu J, Luo T, Fan F. Glioblastoma treatment modalities besides surgery. J Cancer. 2019;10:4793–806.
Daisne J-F, Blumhofer A. Atlas-based automatic segmentation of head and neck organs at risk and nodal target volumes: a clinical validation. Radiat Oncol. 2013;8:154.
Zikou A, Sioka C, Alexiou GA, Fotopoulos A, Voulgaris S, Argyropoulou MI. Radiation necrosis, pseudoprogression, pseudoresponse, and tumor recurrence: imaging challenges for the evaluation of treated gliomas. Contrast Media Mol Imaging. 2018;2018:6828396.
Kessler AT, Bhatt AA. Brain tumour post-treatment imaging and treatment-related complications. Insights Imaging. 2018;9:1057–75.
Hu X, Wong KK, Young GS, Guo L, Wong ST. Support vector machine multiparametric MRI identification of pseudoprogression from tumor recurrence in patients with resected glioblastoma. J Magn Reson Imaging. 2011;33:296–305.
Jang B-S, Jeon SH, Kim IH, Kim IA. Prediction of pseudoprogression versus progression using machine learning algorithm in glioblastoma. Sci Rep. 2018;8:12516.
Kourou K, Exarchos TP, Exarchos KP, Karamouzis MV, Fotiadis DI. Machine learning applications in cancer prognosis and prediction. Comput Struct Biotechnol J. 2014;13:8–17.
Lao J, Chen Y, Li Z-C, Li Q, Zhang J, Liu J, et al. A deep learning-based radiomics model for prediction of survival in glioblastoma multiforme. Sci Rep. 2017;7:10353.
Nie D, Lu J, Zhang H, Adeli E, Wang J, Yu Z, et al. Multi-channel 3D deep feature learning for survival time prediction of brain tumor patients using multi-modal neuroimages. Sci Rep. 2019;9:1103.
Sarkiss CA, Germano IM. Machine learning in neuro-oncology: can data analysis from 5346 patients change decision-making paradigms? World Neurosurg. 2019;124:287–94.
Calabrese E, Villanueva-Meyer JE, Cha S. A fully automated artificial intelligence method for non-invasive, imaging-based identification of genetic alterations in glioblastomas. Sci Rep. 2020;10:11852.
Veronese E, Castellani U, Peruzzo D, Bellani M, Brambilla P. Machine learning approaches: from theory to application in schizophrenia. Comput Math Methods Med. 2013;2013:867924.
Mehdy MM, Ng PY, Shair EF, Saleh NIM, Gomes C. Artificial neural networks in image processing for early detection of breast cancer. Comput Math Methods Med. 2017;2017:2610628.
Chakure A. Random forest classification and its implementation [Internet]. Medium. 2020 [cited 2020 Nov 17]. Available from: https://medium.com/swlh/random-forest-classification-and-its-implementation-d5d840dbead0.
Acknowledgement
We would like to thank the Faculty of Central Inter-Disciplinary Research Facility, Sri Balaji Vidyapeeth for giving us constant support in drafting this review article.
Funding
This review did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
All the authors contributed equally to the conception and design of this review article. TSA: Conceptualization and Critical Evaluation of the work. PSD: Literature search and original draft preparation.
Corresponding author
Ethics declarations
Conflict of interest
The authors Daisy Precilla S and Anitha T.S declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Daisy, P.S., Anitha, T.S. Can artificial intelligence overtake human intelligence on the bumpy road towards glioma therapy?. Med Oncol 38, 53 (2021). https://doi.org/10.1007/s12032-021-01500-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-021-01500-2