Abstract
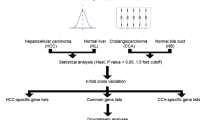
Hepatocellular carcinoma (HCC) is among the commonest kind of malignant tumors, which accounts for more than 500,000 cases of newly diagnosed cancer annually. Many microarray studies for identifying differentially expressed genes (DEGs) in HCC have been conducted, but results have varied across different studies. Here, we performed a meta-analysis of publicly available microarray Gene Expression Omnibus datasets, which covers five independent studies, containing 753 HCC samples and 638 non-tumor liver samples. We identified 192 DEGs that were consistently up-regulated in HCC vs. normal liver tissue. For the 192 up-regulated genes, we performed Kyoto Encyclopedia of Genes and Genomes pathway analysis. To our surprise, besides several cell growth-related pathways, spliceosome pathway was also up-regulated in HCC. For further exploring the relationship between spliceosome pathway and HCC, we investigated the expression data of spliceosome pathway genes in 15 independent studies in Nextbio database (https://www.nextbio.com/b/nextbioCorp.nb). It was found that many genes of spliceosome pathway such as HSPA1A, SNRPE, SF3B2, SF3B4 and TRA2A genes which we identified to be up-regulated in our meta-analysis were generally overexpressed in HCC. At last, using real-time PCR, we also found that BUD31, SF3B2, SF3B4, SNRPE, SPINK1, TPA2A and HSPA1A genes are significantly up-regulated in clinical HCC samples when compared to the corresponding non-tumorous liver tissues. Our study for the first time indicates that many genes of spliceosome pathway are up-regulated in HCC. This finding might put new insights for people’s understanding about the relationship of spliceosome pathway and HCC.



Similar content being viewed by others
References
Jemal A, Bray F, Eard E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90.
Coleman WB. Mechanisms of human hepatocarcinogenesis. Curr Mol Med. 2003;3:573–88.
Jackson PE, Kuang SY, Wang JB, Strickland PT, Munoz A, Kensler TW, Qian GS, Groopman JD. Prospective detection of codon 249 mutations in plasma of hepatocellular carcinoma patients. Carcinogenesis. 2003;24:1657–63.
Kuang SY, Lekawanvijit S, Maneekarn N, Thongsawat S, Brodovicz K, Nelson K, Groopman JD. Hepatitis b 1762t/1764a mutations, hepatitis c infection, and codon 249 p53 mutations in hepatocellular carcinomas from Thailand. Cancer Epidemiol Biomark Prev. 2005;14:380–4.
Law PT, Ching AK, Chan AW, Wong QW, Wong CK, To KF, Wong N. Mir-145 modulates multiple components of the insulin-like growth factor pathway in hepatocellular carcinoma. Carcinogenesis. 2012;33:1134–41.
Ertel A, Verghese A, Byers SW, Ochs M, Tozeren A. Pathway-specific differences between tumor cell lines and normal and tumor tissue cells. Mol Cancer. 2006;5:55.
Hu N, Wang C, Hu Y, Yang HH, Giffen C, Tang ZZ, Han XY, Goldstein AM, Emmert-Buck MR, Buetow KH, Taylor PR, Lee MP. Genome-wide association study in esophageal cancer using genechip mapping 10 k array. Cancer Res. 2005;65:2542–6.
Ramaswamy S, Tamayo P, Rifkin R, Mukherjee S, Yeang CH, Angelo M, Ladd C, Reich M, Latulippe E, Mesirov JP, Poggio T, Gerald W, Loda M, Lander ES, Golub TR. Multiclass cancer diagnosis using tumor gene expression signatures. Proc Natl Acad Sci USA. 2001;98:15149–54.
Nordmann AJ, Kasenda B, Briel M. Meta-analyses: what they can and cannot do. Swiss Med Wkly. 2012;142:w13518.
Chen C, Fu X, Zhang D, Li Y, Xie Y, Li Y, Huang Y. Varied pathways of stage ia lung adenocarcinomas discovered by integrated gene expression analysis. Int J Biol Sci. 2011;7:551–66.
Liu W, Peng Y, Tobin DJ. A new 12-gene diagnostic biomarker signature of melanoma revealed by integrated microarray analysis. PeerJ. 2013;1:e49.
Ramasamy A, Mondry A, Holmes CC, Altman DG. Key issues in conducting a meta-analysis of gene expression microarray datasets. PLoS Med. 2008;5:e184.
Zaravinos A, Lambrou GI, Boulalas I, Delakas D, Spandidos DA. Identification of common differentially expressed genes in urinary bladder cancer. PLoS One. 2011;6:e18135.
Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–5.
Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic rnp machine. Cell. 2009;136:701–18.
Lee JS, Chu IS, Mikaelyan A, Calvisi DF, Heo J, Reddy JK, Thorgeirsson SS. Application of comparative functional genomics to identify best-fit mouse models to study human cancer. Nat Genet. 2004;36:1306–11.
Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF, Mikaelyan A, Roberts LR, Demetris AJ, Sun Z, Nevens F, Roskams T, Thorgeirsson SS. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410–6.
Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70:10202–12.
Roessler S, Long EL, Budhu A, Chen Y, Zhao X, Ji J, Walker R, Jia HL, Ye QH, Qin LX, Tang ZY, He P, Hunter KW, Thorgeirsson SS, Meltzer PS, Wang XW. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology. 2012;142(957–966):e912.
Kim BY, Lee JG, Park S, Ahn JY, Ju YJ, Chung JH, Han CJ, Jeong SH, Yeom YI, Kim S, Lee YS, Kim CM, Eom EM, Lee DH, Choi KY, Cho MH, Suh KS, Choi DW, Lee KH. Feature genes of hepatitis b virus-positive hepatocellular carcinoma, established by its molecular discrimination approach using prediction analysis of microarray. Biochim Biophys Acta. 2004;1739:50–61.
Tung EK, Mak CK, Fatima S, Lo RC, Zhao H, Zhang C, Dai H, Poon RT, Yuen MF, Lai CL, Li JJ, Luk JM, Ng IO. Clinicopathological and prognostic significance of serum and tissue dickkopf-1 levels in human hepatocellular carcinoma. Liver Int. 2011;31:1494–504.
Lamb JR, Zhang C, Xie T, Wang K, Zhang B, Hao K, Chudin E, Fraser HB, Millstein J, Ferguson M, Suver C, Ivanovska I, Scott M, Philippar U, Bansal D, Zhang Z, Burchard J, Smith R, Greenawalt D, Cleary M, Derry J, Loboda A, Watters J, Poon RT, Fan ST, Yeung C, Lee NP, Guinney J, Molony C, Emilsson V, Buser-Doepner C, Zhu J, Friend S, Mao M, Shaw PM, Dai H, Luk JM, Schadt EE. Predictive genes in adjacent normal tissue are preferentially altered by scnv during tumorigenesis in liver cancer and may rate limiting. PLoS One. 2011;6:e20090.
Sung WK, Zheng H, Li S, Chen R, Liu X, Li Y, Lee NP, Lee WH, Ariyaratne PN, Tennakoon C, Mulawadi FH, Wong KF, Liu AM, Poon RT, Fan ST, Chan KL, Gong Z, Hu Y, Lin Z, Wang G, Zhang Q, Barber TD, Chou WC, Aggarwal A, Hao K, Zhou W, Zhang C, Hardwick J, Buser C, Xu J, Kan Z, Dai H, Mao M, Reinhard C, Wang J, Luk JM. Genome-wide survey of recurrent hbv integration in hepatocellular carcinoma. Nat Genet. 2012;44:765–9.
Troyanskaya O, Cantor M, Sherlock G, Brown P, Hastie T, Tibshirani R, Botstein D, Altman RB. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–5.
Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statist Appl Genet Mol Biol. 2004;3:1–25.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodological) 1995:289–300.
da Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13.
Kanehisa M, Goto S. Kegg: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30.
Wu Y, Zuo J, Ji G, Saiyin H, Liu X, Yin F, Cao N, Wen Y, Li JJ, Yu L. Proapoptotic function of integrin beta(3) in human hepatocellular carcinoma cells. Clin Cancer Res 2009: 60–69.
Kupershmid I, Su QJ, Grewal A, Sundaresh S, Halperin I, Flynn J, Shekar M, Wang H, Park J, Cui W, Wall GD, Wisotzkey R, Alag S, Akhtari S, Ronaghi M. Ontology-based meta-analysis of global collections of high-throughput public data. PloS One 2010;5.
Marshall A, Lukk M, Kutter C, Davies S, Alexander G, Odom DT. Global gene expression profiling reveals spink1 as a potential hepatocellular carcinoma marker. PLoS One. 2013;8:e59459.
Nilsen TW. The spliceosome: the most complex macromolecular machine in the cell? BioEssays. 2003;25:1147–9.
Matera AG, Wang Z. A day in the life of the spliceosome. Nat Rev Mol Cell Biol. 2014;15:108–21.
Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. Deciphering the splicing code. Nature. 2010;465:53–9.
Dehm SM. Test-firing ammunition for spliceosome inhibition in cancer. Clin Cancer Res. 2013;19:6064–6.
Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H, Tani T, Horinouchi S, Yoshida M. Spliceostatin a targets sf3b and inhibits both splicing and nuclear retention of pre-mrna. Nat Chem Biol. 2007;3:576–83.
Leu JI, Pimkina J, Pandey P, Murphy ME, George DL. Hsp70 inhibition by the small-molecule 2-phenylethynesulfonamide impairs protein clearance pathways in tumor cells. Molecular Cancer Res MCR. 2011;9:936–47.
Guo F, Rocha K, Bali P, Pranpat M, Fiskus W, Boyapalle S, Kumaraswamy S, Balasis M, Greedy B, Armitage ES, Lawrence N, Bhalla K. Abrogation of heat shock protein 70 induction as a strategy to increase antileukemia activity of heat shock protein 90 inhibitor 17-allylamino-demethoxy geldanamycin. Cancer Res. 2005;65:10536–44.
Quidville V, Alsafadi S, Goubar A, Commo F, Scott V, Pioche-Durieu C, Girault I, Baconnais S, Le Cam E, Lazar V, Delaloge S, Saghatchian M, Pautier P, Morice P, Dessen P, Vagner S, Andre F. Targeting the deregulated spliceosome core machinery in cancer cells triggers mtor blockade and autophagy. Cancer Res. 2013;73:2247–58.
Anchi T, Tamura K, Furihata M, Satake H, Sakoda H, Kawada C, Kamei M, Shimamoto T, Fukuhara H, Fukata S, Ashida S, Karashima T, Yamasaki I, Yasuda M, Kamada M, Inoue K, Shuin T. Snrpe is involved in cell proliferation and progression of high-grade prostate cancer through the regulation of androgen receptor expression. Oncol Lett. 2012;3:264–8.
Best A, Dagliesh C, Ehrmann I, Kheirollahi-Kouhestani M, Tyson-Capper A, Elliott DJ. Expression of tra2 beta in cancer cells as a potential contributory factor to neoplasia and metastasis. Int J Cell Biol. 2013;2013:843781.
Watermann DO, Tang Y, Zur Hausen A, Jager M, Stamm S, Stickeler E. Splicing factor tra2-beta1 is specifically induced in breast cancer and regulates alternative splicing of the cd44 gene. Cancer Res. 2006;66:4774–80.
Gabriel B, Zur Hausen A, Bouda J, Boudova L, Koprivova M, Hirschfeld M, Jager M, Stickeler E. Significance of nuclear htra2-beta1 expression in cervical cancer. Acta Obstet Gynecol Scand. 2009;88:216–21.
Yin J, Zhu JM, Shen XZ. New insights into pre-mrna processing factor 19: a multi-faceted protein in humans. Biol Cell. 2012;104:695–705.
Voglauer R, Chang MW, Dampier B, Wieser M, Baumann K, Sterovsky T, Schreiber M, Katinger H, Grillari J. Snev overexpression extends the life span of human endothelial cells. Exp Cell Res. 2006;312:746–59.
Modrek B, Lee C. A genomic view of alternative splicing. Nat Genet. 2002;30:13–9.
Liu HX, Cartegni L, Zhang MQ, Krainer AR. A mechanism for exon skipping caused by nonsense or missense mutations in brca1 and other genes. Nat Genet. 2001;27:55–8.
Barbour AP, Reeder JA, Walsh MD, Fawcett J, Antalis TM, Gotley DC. Expression of the cd44v2-10 isoform confers a metastatic phenotype: importance of the heparan sulfate attachment site cd44v3. Cancer Res. 2003;63:887–92.
Neklason DW, Solomon CH, Dalton AL, Kuwada SK, Burt RW. Intron 4 mutation in apc gene results in splice defect and attenuated fap phenotype. Fam Cancer. 2004;3:35–40.
Kalnina Z, Zayakin P, Silina K, Line A. Alterations of pre-mrna splicing in cancer. Genes Chromosom Cancer. 2005;42:342–57.
Lim HY, Sohn I, Deng S, Lee J, Jung SH, Mao M, Xu J, Wang K, Shi S, Joh JW, Choi YL, Park CK. Prediction of disease-free survival in hepatocellular carcinoma by gene expression profiling. Ann Surg Oncol. 2013;20:3747–53.
Stefanska B, Huang J, Bhattacharyya B, Suderman M, Hallett M, Han ZG, Szyf M. Definition of the landscape of promoter DNA hypomethylation in liver cancer. Cancer Res. 2011;71:5891–903.
Huang Y, Chen HC, Chiang CW, Yeh CT, Chen SJ, Chou CK. Identification of a two-layer regulatory network of proliferation-related micrornas in hepatoma cells. Nucleic Acids Res. 2012;40:10478–93.
Chuma M, Sakamoto M, Yamazaki K, Ohta T, Ohki M, Asaka M, Hirohashi S. Expression profiling in multistage hepatocarcinogenesis: identification of hsp70 as a molecular marker of early hepatocellular carcinoma. Hepatology. 2003;37:198–207.
Ye QH, Qin LX, Forgues M, He P, Kim JW, Peng AC, Simon R, Li Y, Robles AI, Chen Y, Ma ZC, Wu ZQ, Ye SL, Liu YK, Tang ZY, Wang XW. Predicting hepatitis b virus-positive metastatic hepatocellular carcinomas using gene expression profiling and supervised machine learning. Nat Med. 2003;9:416–23.
Wurmbach E, Chen YB, Khitrov G, Zhang W, Roayaie S, Schwartz M, Fiel I, Thung S, Mazzaferro V, Bruix J, Bottinger E, Friedman S, Waxman S, Llovet JM. Genome-wide molecular profiles of hcv-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45:938–47.
Kaposi-Novak P, Libbrecht L, Woo HG, Lee YH, Sears NC, Coulouarn C, Conner EA, Factor VM, Roskams T, Thorgeirsson SS. Central role of c-myc during malignant conversion in human hepatocarcinogenesis. Cancer Res. 2009;69:2775–82.
Liao YL, Sun YM, Chau GY, Chau YP, Lai TC, Wang JL, Horng JT, Hsiao M, Tsou AP. Identification of sox4 target genes using phylogenetic footprinting-based prediction from expression microarrays suggests that overexpression of sox4 potentiates metastasis in hepatocellular carcinoma. Oncogene. 2008;27:5578–89.
Mas VR, Maluf DG, Archer KJ, Yanek K, Kong X, Kulik L, Freise CE, Olthoff KM, Ghobrial RM, McIver P, Fisher R. Genes involved in viral carcinogenesis and tumor initiation in hepatitis c virus-induced hepatocellular carcinoma. Mol Med. 2009;15:85–94.
Segal E, Sirlin CB, Ooi C, Adler AS, Gollub J, Chen X, Chan BK, Matcuk GR, Barry CT, Chang HY, Kuo MD. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol. 2007;25:675–80.
Acknowledgments
This work was supported by National Natural Science Foundation of China (Grant No. 31071186, to Dr. Cao). Weijin Xu was supported by National Top Talent Undergraduate Training Program (NTTUTP).
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Weijin Xu and Huixing Huang have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xu, W., Huang, H., Yu, L. et al. Meta-analysis of gene expression profiles indicates genes in spliceosome pathway are up-regulated in hepatocellular carcinoma (HCC). Med Oncol 32, 96 (2015). https://doi.org/10.1007/s12032-014-0425-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-014-0425-6