Abstract
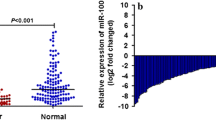
MicroRNA-25 (miR-25) has recently been found to be involved in many critical processes in human malignancies. We aimed to investigate the expression pattern and prognostic role of miR-25 in colorectal cancer. Colorectal cancer and adjacent normal specimens from 186 patients who had not received neoadjuvant chemotherapy were collected. The expression of miR-25 was detected with a quantitative real-time PCR assay, and the association of miR-25 with overall patient survival was analyzed via statistical analysis. The results indicated that the level of miR-25 expression was significantly elevated in colorectal cancer compared with the level observed in the adjacent normal tissue. It was also demonstrated that miR-25 expression is associated with tumor invasion, lymph node metastasis, distant metastasis and the TNM stage of colorectal cancer. In addition, a Kaplan–Meier analysis revealed that an increased level of miR-25 expression is associated with a poor overall survival of patients. A multivariate survival analysis also indicated that miR-25 is an independent prognostic marker after adjusting for known prognostic factors. These results prove that miR-25 expression is increased in colorectal cancer and is associated with tumor progression. This study also provides the first evidence that miR-25 is an independent prognostic factor for patients with colorectal cancer, indicating the potential role of miR-25 as a highly specific and sensitive biomarker.

Similar content being viewed by others
References
Siegel R, Naishadham D, Jemal A. Cancer statistics 2012. CA Cancer J Clin. 2012;62(1):10–29. doi:10.3322/caac.20138.
Center MM, Jemal A, Smith RA, Ward E. Worldwide variations in colorectal cancer. CA Cancer J Clin. 2009;59(6):366–78. doi:10.3322/caac.20038.
Deng SX, An W, Gao J, Yin J, Cai QC, Yang M, et al. Factors influencing diagnosis of colorectal cancer: a hospital-based survey in China. J Dig Dis. 2012;13(10):517–24. doi:10.1111/j.1751-2980.2012.00626.x.
Chen HM, Weng YR, Jiang B, Sheng JQ, Zheng P, Yu CG, et al. Epidemiological study of colorectal adenoma and cancer in symptomatic patients in China between 1990 and 2009. J Dig Dis. 2011;12(5):371–8. doi:10.1111/j.1751-2980.2011.00531.x.
Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, Smith T, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62(4):220–41. doi:10.3322/caac.21149.
Wolpin BM, Meyerhardt JA, Mamon HJ, Mayer RJ. Adjuvant treatment of colorectal cancer. CA Cancer J Clin. 2007;57(3):168–85.
Shukla GC, Singh J, Barik S. MicroRNAs: processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol. 2011;3(3):83–92.
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi:10.1016/j.cell.2009.01.002.
John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2(11):e363. doi:10.1371/journal.pbio.0020363.
Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z, et al. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18(10):1165–78. doi:10.1101/gad.1184704.
Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–66. doi:10.1038/nrc1997.
Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103(7):2257–61. doi:10.1073/pnas.0510565103.
Schee K, Boye K, Abrahamsen TW, Fodstad O, Flatmark K. Clinical relevance of microRNA miR-21, miR-31, miR-92a, miR-101, miR-106a and miR-145 in colorectal cancer. BMC Cancer. 2012;12:505. doi:10.1186/1471-2407-12-505.
Schimanski CC, Frerichs K, Rahman F, Berger M, Lang H, Galle PR, et al. High miR-196a levels promote the oncogenic phenotype of colorectal cancer cells. World J Gastroenterol WJG. 2009;15(17):2089–96.
Bandres E, Cubedo E, Agirre X, Malumbres R, Zarate R, Ramirez N, et al. Identification by real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29. doi:10.1186/1476-4598-5-29.
Motoyama K, Inoue H, Takatsuno Y, Tanaka F, Mimori K, Uetake H, et al. Over- and under-expressed microRNAs in human colorectal cancer. Int J Oncol. 2009;34(4):1069–75.
Nie J, Liu L, Zheng W, Chen L, Wu X, Xu Y, et al. microRNA-365, down-regulated in colon cancer, inhibits cell cycle progression and promotes apoptosis of colon cancer cells by probably targeting Cyclin D1 and Bcl-2. Carcinogenesis. 2012;33(1):220–5. doi:10.1093/carcin/bgr245.
Liu L, Chen L, Xu Y, Li R, Du X. microRNA-195 promotes apoptosis and suppresses tumorigenicity of human colorectal cancer cells. Biochem Biophys Res Commun. 2010;400(2):236–40. doi:10.1016/j.bbrc.2010.08.046.
Poliseno L, Salmena L, Riccardi L, Fornari A, Song MS, Hobbs RM, et al. Identification of the miR-106b ~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal. 2010;3(117):ra29. doi:10.1126/scisignal.2000594.
Birks DK, Barton VN, Donson AM, Handler MH, Vibhakar R, Foreman NK. Survey of MicroRNA expression in pediatric brain tumors. Pediatr Blood Cancer. 2011;56(2):211–6. doi:10.1002/pbc.22723.
Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13(3):272–86. doi:10.1016/j.ccr.2008.02.013.
Dacic S, Kelly L, Shuai Y, Nikiforova MN. miRNA expression profiling of lung adenocarcinomas: correlation with mutational status. Mod Pathol. 2010;23(12):1577–82. doi:10.1038/modpathol.2010.152.
Li Y, Tan W, Neo TW, Aung MO, Wasser S, Lim SG, et al. Role of the miR-106b-25 microRNA cluster in hepatocellular carcinoma. Cancer Sci. 2009;100(7):1234–42. doi:10.1111/j.1349-7006.2009.01164.x.
Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–74. doi:10.1038/nrg3074.
He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–31. doi:10.1038/nrg1379.
Mendell JT. MicroRNAs: critical regulators of development, cellular physiology and malignancy. Cell Cycle. 2005;4(9):1179–84.
Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11(12):849–64. doi:10.1038/nrc3166.
Pogribny IP, Tryndyak VP, Boyko A, Rodriguez-Juarez R, Beland FA, Kovalchuk O. Induction of microRNAome deregulation in rat liver by long-term tamoxifen exposure. Mutat Res. 2007;619(1–2):30–7. doi:10.1016/j.mrfmmm.2006.12.006.
Zhao H, Wang Y, Yang L, Jiang R, Li W. miR-25 promotes gastric cancer cells growth and motility by targeting RECK. Mol Cell Biochem. 2013;. doi:10.1007/s11010-013-1829-x.
Zhang H, Zuo Z, Lu X, Wang L, Wang H, Zhu Z. MiR-25 regulates apoptosis by targeting Bim in human ovarian cancer. Oncol Rep. 2012;27(2):594–8. doi:10.3892/or 2011.1530.
Xu JY, Yang LL, Ma C, Huang YL, Zhu GX, Chen QL. MiR-25-3p attenuates the proliferation of tongue squamous cell carcinoma cell line Tca8113. Asian Pac J Trop Biomed. 2013;6(9):743–7. doi:10.1016/S1995-7645(13)60130-3.
Wang Y, Li Z, He C, Wang D, Yuan X, Chen J, et al. MicroRNAs expression signatures are associated with lineage and survival in acute leukemias. Blood Cells Mol Dis. 2010;44(3):191–7. doi:10.1016/j.bcmd.2009.12.010.
Xu X, Chen Z, Zhao X, Wang J, Ding D, Wang Z, et al. MicroRNA-25 promotes cell migration and invasion in esophageal squamous cell carcinoma. Biochem Biophys Res Commun. 2012;421(4):640–5. doi:10.1016/j.bbrc.2012.03.048.
Conflict of interests
None.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Li, X., Yang, C., Wang, X. et al. The expression of miR-25 is increased in colorectal cancer and is associated with patient prognosis. Med Oncol 31, 781 (2014). https://doi.org/10.1007/s12032-013-0781-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-013-0781-7