Abstract
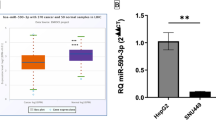
MicroRNAs (miRNAs) are an abundant class of short noncoding RNAs that can posttranscriptionally regulate gene expression in animals. They are also involved in cancer initiation and progression, and their expression profiles serve as phenotypic signatures of different cancers. The roles played by microRNAs specifically in “micromanagement of metastasis” has been addressed only recently. The molecular mechanisms of hepatocellular carcinoma (HCC) metastasis are still poorly understood. Recent evidence implies genetic determinants of cancer metastasis. Because gene expression signature significantly differs between primary metastasis-free HCC and primary HCC with intrahepatic metastases, miRNA expression in those primary HCC may change correspondingly. The 28 up-regulated miRNAs, part of the reported miRNA profiles of HCC, were compared in primary HCC with or without metastases. Only eight miRNAs were found to be significantly up-regulated in primary HCC with metastases while miR-9 had the highest hold change. miR-9 was highly expressed in SK-Hep-1 cell when compared with other hepatoma cell lines and downregulation of miR-9 reduced SK-Hep-1 cell invasion. E-cadherin, a tumor invasion suppressor in HCC, was found to be a putative gene target of miR-9. E-cadherin was up-regulated by miR-9 inhibitor. The findings suggest miR-9 could be involved in HCC metastasis.




Similar content being viewed by others
References
Toyosaka A, Okamoto E, Mitsunobu M, et al. Intrahepatic metastases in hepatocellular carcinoma: evidence for spread via the portal vein as an efferent vessel. Am J Gastroenterol. 1996;91:1610–5.
Nguyen DX, Massague J. Genetic determinants of cancer metastasis. Nat Rev Genet. 2007;8:341–52. doi:10.1038/nrg2101.
Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi:10.1038/nrc1997.
Steeg PS. Cancer: micromanagement of metastasis. Nature. 2007;449:671–3. doi:10.1038/449671a.
Ma L, Weinberg RA. Micromanagers of malignancy: role of microRNAs in regulating metastasis. Trends Genet. 2008;24:448–56. doi:10.1016/j.tig.2008.06.004.
Ye QH, Qin LX, Forgues M, et al. Predicting hepatitis B virus-positive metastatic hepatocellular carcinomas using gene expression profiling and supervised machine learning. Nat Med. 2003;9:416–23. doi:10.1038/nm843.
Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi:10.1038/nature03702.
Ladeiro Y, Couchy G, Balabaud C, et al. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955–63. doi:10.1002/hep.22256.
Wang Y, Lee AT, Ma JZ, et al. Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J Biol Chem. 2008;283:13205–15. doi:10.1074/jbc.M707629200.
Budhu A, Jia HL, Forgues M, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi:10.1002/hep.22160.
Li W, Xie L, He X, et al. Diagnostic and prognostic implications of microRNAs in human hepatocellular carcinoma. Int J Cancer. 2008;123:1616–22. doi:10.1002/ijc.23693.
Murakami Y, Yasuda T, Saigo K, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–45. doi:10.1038/sj.onc.1209283.
Edmondson HA, Steiner PE. Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer. 1954;7:462–503. doi:10.1002/1097-0142(195405)7:3<462::AID-CNCR2820070308>3.0.CO;2-E.
AJCC. AJCC cancer staging manual. New York: Springer; 2002.
Krek A, Grun D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi:10.1038/ng1536.
Lewis BP, Shih IH, Jones-Rhoades MW, et al. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi:10.1016/S0092-8674(03)01018-3.
Huang GT, Lee HS, Chen CH, et al. Correlation of E-cadherin expression and recurrence of hepatocellular carcinoma. Hepatogastroenterology. 1999;46:1923–7.
Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med. 2006;12:895–904. doi:10.1038/nm1469.
Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi:10.1016/j.cell.2004.12.035.
Ma L, Weinberg RA. Micromanagers of malignancy: role of microRNAs in regulating metastasis. Trends Genet. 2008;24(9):448–56.
Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–8. doi:10.1038/nature06174.
Lagos-Quintana M, Rauhut R, Yalcin A, et al. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–9. doi:10.1016/S0960-9822(02)00809-6.
Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi:10.1158/0008-5472.CAN-05-1783.
Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98. doi:10.1016/j.ccr.2006.01.025.
Laios A, O’Toole S, Flavin R, et al. Potential role of miR-9 and miR-223 in recurrent ovarian cancer. Mol Cancer. 2008;7:35. doi:10.1186/1476-4598-1187-1135.
Nass D, Rosenwald S, Meiri E et al. MiR-92b and miR-9/9* are specifically expressed in brain primary tumors and can be used to differentiate primary from metastatic brain tumors. Brain Pathol. 2008; 2: doi:10.1111/j.1750-3639.2008.00184.x.
Wijnhoven BP, Dinjens WN, Pignatelli M. E-cadherin-catenin cell-cell adhesion complex and human cancer. Br J Surg. 2000;87:992–1005. doi:10.1046/j.1365-2168.2000.01513.x.
Acknowledgments
This study was supported by Project of National Natural Sciences Foundation of China (2004Z2-E0132).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hao-Xiang, T., Qian, W., Lian-Zhou, C. et al. MicroRNA-9 reduces cell invasion and E-cadherin secretion in SK-Hep-1 cell. Med Oncol 27, 654–660 (2010). https://doi.org/10.1007/s12032-009-9264-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12032-009-9264-2