Abstract
EBV (Epstein-Barr Virus) and other human DNA viruses are associated with autoimmune syndromes in epidemiologic studies. In this work, immunoglobulin G response to EBV-encoded proteins which share regions with human immune response proteins from the human host including ZEBRA (BZLF-1 encoded protein), BALF-2 recombinase expressed primarily during the viral lytic replication cycle, and EBNA-1 (Epstein-Barr Virus Nuclear Antigen) expressed during the viral latency cycle respectively were characterized using a laser-printed micro-array (PEPperprint.com). IgG response to conserved “A/T hooks” in EBV-encoded proteins such as EBNA-1 and the BALF-2 recombinase related to host DNA-binding proteins including RAG-1 recombinase and histones, and EBV-encoded virokines such as the IL-10 homologue BCRF-1 suggest further directions for clinical research. The author suggests that proteomic “molecular fingerprints” of the immune response to viral proteins shared with human immune response genes are potentially useful in early diagnosis and monitoring of autoantibody production and response to therapy in EBV-related autoimmune syndromes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.

Introduction
DNA viruses such as EBV (Epstein-Barr Virus) have been associated with numerous autoimmune syndromes in epidemiologic studies, but the specific role of infection remains unresolved [1,2,3]. Conventional serologic testing for EBV is available using specific immunoglobulin G (IgG) binding to viral proteins such as EBNA-1 (Epstein-Barr Virus Nuclear Antigen-1), VCA (Viral Capsid Antigen), and EA (viral components termed Early Antigen). A correlation between antigen-specific IgG levels to specific EBV proteins has been reported in SLE, MS (multiple sclerosis), and other autoimmune syndromes [4,5,6,7,8,9]. Since the vast majority of the adult population is positive for EBV, it would be useful to have additional laboratory analysis for clinical evaluation and monitoring of autoimmune disease in patients based on response to past EBV infection [10,11,12,13].
Recent developments in proteomic analysis and technology have dramatically increased the power and lowered the cost of many immunological tests [14]. Presently, the standard of diagnosis for many autoimmune diseases is based on ANA (anti-nuclear antibody) assays in which immunoglobulin binding is characterized against the cell cytoplasm and nucleus and related IgG binding assays. While ANA and immunoassay technology can in principle be automated, a more recent approach is the use of specific antigenic host and viral proteins printed in a microarray with sensitivity similar or great than that of the ANA [15, 16]. Using proteomic technology, hundreds or even thousands of epitopes can be analyzed simultaneously with a single serum sample. Detailed mapping of autoreactive viral epitopes could suggest epitopes capable of differentiating healthy patients versus patients with autoimmune syndromes such as SLE and scleroderma in which EBV reactivation, EBNA-1, and ZEBRA expression are evident [17,18,19].
Viral proteins trigger autoreactive IgG is “molecular mimicry” in which short regions of similarity between viral and host proteins are proposed to cross-react and through “epitope spreading” lead to IgG against host proteins in the presence of viral inflammation and defects in host suppressor cells [20,21,22,23,24]. EBV-encoded proteins such as ZEBRA (BZLF-1 protein) share regions with host transcription factors in the fos/jun family and also to host ankyrin proteins that anchor the cytoskeleton and regulate host transcription factors such as p53 and NF-kB important in the immune response [25]. A well-characterized example of “molecular mimicry” between EBV and host protein is the similarity between regions of EBNA-1 and the “Smith Antigen” in SLE [26, 27]. EBNA-1 and other proteins including the viral encoded recombinase BALF-2 protein share a DNA-binding domain termed an A/T hook with both vertebrate and invertebrate transcription factors and DNA-binding proteins such as histones and recombinases [28]. Chronic viruses also encode “virokines” similar to host cytokines [29].
It would therefore be useful to have a sensitive and inexpensive method to generate a “molecular fingerprint” of IgG binding to shared regions of viral and host proteins to compare between healthy patients with autoimmune disease or at risk in addition to available more labor intensive and costly technology such as the ANA. Using “PEPperprint” technology, the authors characterized peptide epitopes that might provide a sensitive and inexpensive method to generate a proteomic “molecular fingerprint” of IgG binding to shared regions of viral and host proteins [30]. Proteomic technology appears to be highly flexible and inexpensive relative to previous whole protein-based technologies and also permits localization of binding to precise intervals of the viral proteins. A limitation of the technology is that some epitopes which require complex folding and tertiary interactions of viral proteins or secondary modifications evident in ELISA binding assays will not be detected [12, 31]. Conclusions regarding clinical utility therefore must await confirmation in additional studies based on direct comparison with other diagnostic systems [14].
Methods
Molecular images of “A/T hook” DNA-binding domains in figures shown were generated by the authors from published public access coordinates of X-ray crystallography coordinates from crystallized RAG-1 nonamer (pdb file 3GNA) and Herpes Simplex ICP8 (pdb file 1URJ). The “pymol” program was used through license from the author. Immunoglobulin G response to immunologically important viral proteins containing regions shared with human immune response proteins were characterized. Serum from a healthy EBV-positive donor was obtained with consent for research as part of an ongoing research study of SLE and scleroderma [32, 33].
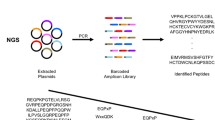
Using proprietary technology (PEPerprint.com, Heidelberg, Germany), overlapping arrays of 15 amino acid long peptides were synthesized on a microchip [30]. Each peptide was progressively printed across the region of interest with 13 amino acid overlap between each peptide or a progression of two amino acids per peptide. Chips were incubated with serum, developed, and analyzed with antibodies specific for immunoglobulin G. Analysis of the immunoglobulin binding was provided by the chip manufacturer (PEPperprint.com) and data was presented using 1 to 100 and 1 to 500 dilutions of serum. Results are presented for both serum dilutions.
In the figures presented in this work, the results of IgG binding are presented in two columns to the right of the printed epitopes, the left column presents the results from the 1/500 serum dilution and the right column presents the results from the 1/100 serum dilution to which an arbitrary factor of “2000” has been added to assist in graphic analysis of results. Results shown in this work suggested that both dilutions of serum gave similar results. Results of IgG binding to shared epitopes were also determined in a patient with an EBV-related autoimmune syndrome, scleroderma, and were significantly different than the healthy control, for example elevated IgG response to BZLF-1 associated epitopes versus EBNA-1 epitopes relative to the healthy control as might be expected due to increased lytic viral replication; however, a complete summary of responses in healthy patients versus autoimmune disease patients is beyond the scope of this work and will be reported elsewhere.
In the tables shown in this work, the actual sequence of each printed peptide is shown with the corresponding location on the chip to the left of the peptide, followed to the right by the actual peptide sequence, followed to the right by the corrected binding intensity donor IgG at 1/500 and 1/100 dilutions of serum. In each 15 amino acid sequence, the approximate position of amino acids with maximal IgG detected binding is underlined. A similar profile or “fingerprint” of response to defined viral peptides could be defined for patients with EBV-related syndromes as well as syndromes associated with reactivation of other herpes viruses such as herpes simplex (an alpha herpes virus latent in neuronal tissues and CMV) or HHV-6 (beta herpes virus latent in hematopoietic stem cells).
Results
IgG response to ZEBRA protein
EBV-encoded ZEBRA protein is a component of the molecular switch between latency and lytic cycles, and IgG response to ZEBRA is present in many autoimmune syndromes. For example, a high level of ZEBRA protein expression is also evident in SLE and scleroderma [32, 33]. As shown in Fig. 1, ZEBRA has a modular structure with three regions each shared with a different host immune response protein. The amino terminus of zebra is a transcription activating domain similar to host immune response transcription activating domains, the central region is a DNA-binding domain similar to the fos/jun DNA-binding proteins that regulate the host immune response, and the terminal region is related to host anchoring proteins regulating NF-kB and innate immune activation, resulting in potential antigen sharing with multiple host DNA-binding proteins including the p53 tumor suppressor [34, 35].
IgG binding in a healthy EBV-positive donor to the entire approximately 220 amino acid long ZEBRA amino acid sequence was analyzed using a laser-printed peptide chip using most the laboratory strain B95-8 and a more virulent human strain of EBV termed “Akata.” Data are shown for the Akata-based peptides, similar or identical to those obtained with B95-8 peptides (B95-8 data not shown). As shown in Table 1, immunoglobulin G response in a healthy donor was confined to two epitopes both located in the amino terminal response of the ZEBRA protein. No immunoglobulin G response was evident to the DNA-binding or ZANK (Zebra ANKyrin-like regions) of zebra in either B95-8 or Akata protein sequences suggesting that this region of ZEBRA is not the source of autoantibodies to p53 and related protein in SLE [36, 37]. Similar IgG binding epitopes in ZEBRA were evident in a patient with scleroderma as will be discussed in more detail elsewhere (data not shown). IgG binding regions of ZEBRA could be used as a part of a molecular fingerprint of IgG binding to regions of EBV shared with host genes (see discussion).
IgG response to EBNA-1 peptide regions shared with host Smith antigen and P53/TRAF binding domains
EBNA-1 protein is the primary protein expressed during EBV latency and is highly antigenic for the lifetime of the host due to periodic viral reactivation and cell lysis releasing small amounts of EBNA-1 into the circulation [38]. Despite the nuclear localization of EBNA-1 as suggested by the name “Epstein-Barr virus nuclear antigen,” the EBNA-1 generates a persistent immunoglobulin G response in both healthy patients and patients with autoimmune syndromes. The unique role of EBNA-1 in viral latency and persistence has decades of research on both the humoral and cellular immune response to EBNA-1 as a tool for viral diagnosis and potentially a tool for antiviral therapy and vaccines [7, 17, 19]. Because of the highly repetitive sequences of certain EBNA-1 regions and the large size of the protein, only specific defined regions of EBNA-1 were analyzed in this work, in contrast to the entire amino acid sequence of ZEBRA protein from two different EBV strains (Table 1).
Research has confirmed that suppressor mechanisms in healthy patients normally limit “epitope spreading” the similar regions of EBNA-1 and host antigens [26, 27, 39]. Several decades ago, molecular mimicry between EBNA-1 and the SLE Smith antigen was observed and subsequently validated in animal models as a mechanism of viral pathogenesis. To facilitate comparison with previous studies, the PEPperprint microchip was applied to a well-characterized EBNA-1 protein epitope similar to the Smith antigen (Table 2). As shown in Table 3, another region of EBNA-1 unrelated to the Smith antigen-like regions that binds to host transcription factors P53 and TRAF also generated a significant host IgG response [40]. This region of EBNA-1 is not related to ankyrin-like regions of ZEBRA and is also unlike regions of p53 binding autoantibodies described in SLE and related syndromes [36, 37].
As with the ZEBRA protein regions, specific IgG binding epitopes of EBNA-1 protein might be a useful diagnostic marker for characterizing a normal response to EBV infection and also for identifying defects in immune suppression in a variety of EBV-related syndromes. Remarkably, a peak of IgG binding was evident overlapping the EBNA-1 smith antigen-like sequence, which also co-localizes to a longer peptide: “PPRRPPPGRRPFFHPVGEADYFEYHQE (EBNA-1 391-420)” previously shown by ELISA assay to correlate with diagnosis of MS [18]. Similar results correlating IgG binding and also T lymphocyte response progression of MS and ELISA using EBNA-1 peptide region 400–641 containing the Smith Ag like region with both T lymphocyte and IgG binding were reported by another MS case–control study [7, 17, 19]. If in fact the specific epitope in EBNA-1 correlating with progression in MS is similar or identical to the EBV associated “Smith antigen” identified in EBV as an early antigen in SLE, this observation might suggest that “proteomic footprinting” can identify previously unknown relationships between pathogenesis and prognosis of MS and SLE (see “Discussion”).
IgG response to A/T hook region peptides of EBNA-1 shared with viral recombinases and host RAG-1 recombinase
The “A/T hook” domain is a family of conserved DNA-binding proteins that bind A/T-rich regions of DNA such as the immunoglobulin nonamer region (Fig. 2). A/T hook domains are also present in host histone proteins as well as both vertebrate and invertebrate DNA-binding proteins including the RAG-1 recombinase [41, 42]. The vertebrate RAG-1 protein is required for generation of the host immune repertoire and binds A/T-rich regions of immunoglobulin and T cell receptor genes with an A/T hook domain shared with a conserved herpes virus recombinase [43]. Two different amino terminal EBNA-1 A/T hook regions are required for viral replication during genome latency probably by linking the host DNA replication and transcription process to specific regions of the viral episome [28]. Comparison between the EBNA-1 A/T hook 1 region of RAG-1, EBNA-1, EBV-encoded BALF-2, and of other BALF-2 like proteins in herpes simplex (ICP8) and CMV and EBV (BALF-2) are shown in Fig. 2. The crystal structure of herpes simplex ICP8 A/T hook is similar to the RAG-1 A/T hook, although little primary amino acid homology is evident (Fig. 3).
Amino acid similarities of A/T hook regions in EBNA-1-1, RAG-1, and herpes virus recombinases. Top panel: An A/T hook region IgG-binding epitope in the amino terminus of EBNA-1 protein is similar in amino acid sequence a conserved region of both the RAG-1 A/T hook and to widely distributed invertebrate A/T hook proteins hin and engrailed. Bottom panel: Herpes viruses also contain an A/T hook (Swiss-Prot accession numbers provided for ICP8 from herpes simplex, CMV, and EBV BALF-2). Viral A/T hooks are significantly different in primary amino acid sequence but have a similar antigenic structure and thus could be useful in quantifying host response both to herpes viruses and related host proteins such as RAG-1 and histones
A/T hook domain from host RAG-1 and herpes simplex virus ICP8. IgG-binding regions of herpes ICP-8 and corresponding regions of RAG-1 are shown (box). The human RAG-1 protein (recombination activating gene encoded protein) A/T hook has been crystallized bound to the A/T-rich nonamer region of immunoglobulin and T cell receptor genes, and the corresponding region of the herpes simplex ICP-8 protein has also been crystallized although not bound to DNA. Very similar structures are evident in A/T hook proteins despite divergent amino acid sequences
As shown in Fig. 3, the A/T hook is exposed on the surface of DNA-binding proteins (published data from host RAG-1 and unpublished data derived from viral ICP8 proteins shown) and therefore would presumably provoke a strong immunoglobulin response if presented to the host immune system. As shown in Table 4, a high level of immunoglobulin G response is present to the first A/T hook region of EBNA-1 and a significantly lower response to the second hook region of EBNA-1. The difference in response to these two similar EBNA-1 structural regions thus specificity is present favoring recognition of the first EBNA-1 hook region over the second hook region despite similar protein tertiary structure. Preliminary data suggests that a much lower response A/T hook regions of EBNA-1 is present in a patient with scleroderma relative to the data shown from a healthy control patient, suggesting that A/T hook regions could be useful to categorize normal and abnormal response to EBNA-1 (unpublished observations).
A/T hook regions described could provide a molecular fingerprint for diagnosis and evaluation of autoimmune syndromes as well as a specific marker for previous EBV and related herpes virus infection. As shown in Table 5, the A/T hook region of herpes simplex ICP8 protein is highly antigenic in a healthy donor. IgG response to the BALF-2 A/T hook and host RAG-1 protein was present but at a much decreased level to the A/T hooks of EBNA-1 or ICP-8 (data not shown). The authors suggest that in pathologic inflammation, EBNA-1 and related virus A/T hooks could serve as autoantigens, since the A/T hook is highly immunogenic in EBNA-1 and other herpes viruses.
IgG response to EBV-encoded “virokine” BCRF-1-encoded protein
Many viral pathogens including EBV encode cytokine like molecules termed “virokines” and also viral-encoded cytokine receptors [44,45,46]. Virokines may also be reduced or masked as targets of the host immune response due to their small size and secondary modifications such as carboxylation [29]. Virokines could interfere with host-encoded cytokines through a variety of mechanisms. Preliminary results obtained with a healthy EBV-positive donor and also a patient with scleroderma, an EBV-associated autoimmune disease, suggest that most of the IgG response against virokine IL-10 is a very low level and restricted primarily to regions of the BCRF-1 protein that are identical to the host IL-10 (data not shown). These preliminary results are consistent with previous studies demonstrating that most of the IgG response to virokines is directed at shared epitopes between virokine and host cytokine (Fig. 4).
Virokines and viral-encoded cytokine receptors as targets for host IgG response. A schematic diagram illustrates several mechanisms through which host IgG against viral proteins similar to host cytokines (virokines) and cytokine receptors could interfere with endogenous regulation of suppressor cell regulatory cytokines such as IL-10. EBV-encoded IL-10 like BCRF-1 is approximately 90% identical to host IL-10 and preliminary results suggest that the immune response to BCRF-1-encoded protein is extremely low and restricted to areas of homology to the host IL-10 cytokine (see “Discussion”)
If further studies confirm that virokines such as the BCL-2-encoded EBV protein are poorly antigenic due to the extensive sharing of epitopes with host cytokines, then the host would be vulnerable to the partial agonist and partial antagonist properties of the EBV-encoded proteins such as BCRF-1 acting on the IL-10 receptor, and this could suggest a paradigm for the pathogenic effects of other shared genes [47,48,49,50,51]. In addition, current studies that characterize levels of IL-10 in autoimmune syndromes may in fact be measuring a combination of viral-encoded virokines and host cytokines. The authors suggest that characterization of IgG response to virokines, viral-encoded cytokine receptors, and host cytokines such as IL-10 is a novel use of proteomic molecular fingerprinting that could also help to distinguish between healthy controls and patients with autoimmune syndromes.
Discussion
Our preliminary results with defined IgG-binding regions of the ZEBRA, EBNA-1, and virokine BCRF-1 protein in a healthy donor suggest that proteomic “molecular fingerprinting” tool significantly lowers the cost and time required for analysis. Further population-based studies would be useful to compare between healthy patients with autoimmune disease or at risk in addition to available more labor-intensive and costly technology such as the ANA. Proteomic assays could potentially be used both for diagnosis and also to monitor response to therapy. In particular, using inexpensive and highly automated “molecular fingerprints,” it might be possible in the future to identify patients at risk of autoimmune syndromes prior to development of symptoms based solely on their response to specific epitopes in viral proteins and shared host proteins. Targeting therapy prior to onset of disease would limit disease progression and therapy-related adverse events.
Published data from an FDA-approved protein micro-array showed that levels of IgG response to host proteins and EBV-encoded EBNA-1 protein and other viral proteins could diagnosis both SLE and scleroderma with sensitivity and specificity similar or greater to conventional ANA testing [15]. Addition of viral protein antigens in this assay significantly improved the sensitivity and specificity, although the specific IgG-binding epitopes recognized by immunoglobulin G were not disclosed [15, 16]. In this work, IgG-binding results are presented derived from a proprietary laser-printed peptide microchip containing overlapping peptides from immunologically important EBV proteins to illustrate the potential of this new technology.
While abnormal immune response to EBV-specific proteins such as EBNA-1 has been described in autoimmune syndromes, specific short epitopes described in the work could facilitate proteomic “molecular fingerprints in autoimmune disease” [7, 17,18,19]. The A/T hook region of EBNA-1 and many herpes virus and host proteins as well as short IgG binding regions in the lytic ZEBRA switch protein described in this work also appear to be a promising antigen for “molecular footprints” of other inflammatory and autoimmune syndromes. Viral proteins such as the viral recombinase BALF-2 protein in Epstein-Barr virus and its homolog ICP8 in herpes simplex and other alpha, beta, and gamma Herpesviridae could also provide epitopes defining parameters such as viral reactivation and molecular mimicry with host proteins [25, 43]. The A/T hook regions of viral and host proteins are shared between both eukaryotic and prokaryotic DNA-binding proteins such as the RAG-1 recombinase and as histones in both vertebrates and invertebrates including the developmentally regulated “hin” and “engrailed” families [41].
Other authors have also recently noted significant overlap between proteins in the human immune response expressed by papilloma virus, another DNA virus correlated with SLE and other human autoimmune syndromes [47,48,49,50,51]. Thus, it seems that “gene sharing” between host immune response genes is not limited to transcription factors and virokines but may include all aspects of the host immune response. Preliminary data obtained from the immune response to some shared gene-encoded proteins reported in this work suggests that while some shared gene-encoded proteins such as BZLF-1 and EBNA-1 are highly antigenic and thus trigger IgG against self-proteins, other EBV-encoded shared genes such as virokines may be poorly antigenic, permitting the viral-encoded projects to function as antagonists or partial agonists of the host immune response. The author suggests that relatively inexpensive new technology such as “proteomic fingerprinting” may be useful to define differences in immune response between patients with autoimmune syndromes and healthy controls including immune response to “shared genes.”
Change history
26 November 2019
The below funding information was missing in the published article.
References
Dreyfus DH. Autoimmune disease: a role for new anti-viral therapies? Autoimmun Rev. 2011;11(2):88–97.
Farina A, Farina GA. Fresh insights into disease etiology and the role of microbial pathogens. Curr Rheumatol Rep. 2015;18(1):1.
Shoenfeld Y. Everything is autoimmune until proven otherwise. Clin Rev Allergy Immunol. 2013;45(2):149–51.
Csuka D, Banati M, Rozsa C, Fust G, Illes Z. High anti-EBNA-1 IgG levels are associated with early-onset myasthenia gravis. Eur J Neurol. 2012;19(6):842–6.
Csuka D, Varga L, Farkas H, Fust G. Strong correlation of high EBNA-1-IgG levels with edematous attacks involving upper airway mucosa in hereditary angioedema due to C1-inhibitor deficiency. Mol Immunol. 2012;49(4):649–54.
Lassmann H, Niedobitek G, Aloisi F, Middeldorp JM, NeuroproMiSe EBVWG. Epstein-Barr virus in the multiple sclerosis brain: a controversial issue—report on a focused workshop held in the Centre for Brain Research of the Medical University of Vienna, Austria. Brain. 2011;134(Pt 9):2772–86.
Lunemann JD, Tintore M, Messmer B, Strowig T, Rovira A, Perkal H, et al. Elevated Epstein-Barr virus-encoded nuclear antigen-1 immune responses predict conversion to multiple sclerosis. Ann Neurol. 2010;67(2):159–69.
Mameli G, Cossu D, Cocco E, Masala S, Frau J, Marrosu MG, et al. EBNA-1 IgG titers in Sardinian multiple sclerosis patients and controls. J Neuroimmunol. 2013;264(1–2):120–2.
Sundar K, Jacques S, Gottlieb P, Villars R, Benito ME, Taylor DK, et al. Expression of the Epstein-Barr virus nuclear antigen-1 (EBNA-1) in the mouse can elicit the production of anti-dsDNA and anti-Sm antibodies. J Autoimmun. 2004;23(2):127–40.
Ingram G, Bugert JJ, Loveless S, Robertson NP. Anti-EBNA-1 IgG is not a reliable marker of multiple sclerosis clinical disease activity. Eur J Neurol. 2010;17(11):1386–9.
Middeldorp JM, Herbrink P. Epstein-Barr virus specific marker molecules for early diagnosis of infectious mononucleosis. J Virol Methods. 1988;21(1–4):133–46.
Schenk BI, Michel PO, Enders G, Thilo N, Radtke M, Oker-Blom C, et al. Evaluation of a new ELISA for the detection of specific IgG to the Epstein-Barr nuclear antigen 1 (EBNA-1). Clin Lab. 2007;53(3–4):151–5.
Yadav P, Carr MT, Yu R, Mumbey-Wafula A, Spatz LA. Mapping an epitope in EBNA-1 that is recognized by monoclonal antibodies to EBNA-1 that cross-react with dsDNA. Immun Inflamm Dis. 2016;4(3):362–75.
Davies JM, Mackay IR, Rowley MJ. Rheumatoid arthritis sera react with a phage-displayed peptide selected by a monoclonal antibody to type II collagen that has homology to EBNA-1. Autoimmunity. 1999;30(1):53–9.
Fattal I, Shental N, Molad Y, Gabrielli A, Pokroy-Shapira E, Oren S, et al. Epstein-Barr virus antibodies mark systemic lupus erythematosus and scleroderma patients negative for anti-DNA. Immunology. 2014;141(2):276–85.
Putterman C, Wu A, Reiner-Benaim A, Batty DS Jr, Sanz I, Oates J, et al. SLE-key((R)) rule-out serologic test for excluding the diagnosis of systemic lupus erythematosus: developing the ImmunArray iCHIP(®). J Immunol Methods. 2016;429:1–6.
Comabella M, Montalban X, Horga A, Messmer B, Kakalacheva K, Strowig T, et al. Antiviral immune response in patients with multiple sclerosis and healthy siblings. Mult Scler. 2010;16(3):355–8.
Jafari N, van Nierop GP, Verjans GM, Osterhaus AD, Middeldorp JM, Hintzen RQ. No evidence for intrathecal IgG synthesis to Epstein Barr virus nuclear antigen-1 in multiple sclerosis. J Clin Virol. 2010;49(1):26–31.
Kakalacheva K, Regenass S, Wiesmayr S, Azzi T, Berger C, Dale RC, Brilot F, Münz C, Rostasy K, Nadal D, Lünemann J Infectious mononucleosis triggers generation of IgG auto-antibodies against native myelin oligodendrocyte glycoprotein. Viruses 2016;8(2).
Mackay IR, Leskovsek NV, Rose NR. The odd couple: a fresh look at autoimmunity and immunodeficiency. J Autoimmun. 2010;35(3):199–205.
Rose NR. Molecular mimicry and clonal deletion: a fresh look. J Theor Biol. 2015;375:71–6.
Wucherpfennig KW. Structural basis of molecular mimicry. J Autoimmun. 2001;16(3):293–302.
Yadav P, Tran H, Ebegbe R, Gottlieb P, Wei H, Lewis RH, et al. Antibodies elicited in response to EBNA-1 may cross-react with dsDNA. PLoS One. 2011;6(1):e14488.
Yaniv G, Twig G, Shor DB, Furer A, Sherer Y, Mozes O, et al. A volcanic explosion of autoantibodies in systemic lupus erythematosus: a diversity of 180 different antibodies found in SLE patients. Autoimmun Rev. 2015;14(1):75–9.
Dreyfus DH. Gene sharing between Epstein-Barr virus and human immune response genes. Immunol Res. 2017;65(1):37–45.
Poole BD, Gross T, Maier S, Harley JB, James JA. Lupus-like autoantibody development in rabbits and mice after immunization with EBNA-1 fragments. J Autoimmun. 2008;31(4):362–71.
Poole BD, Templeton AK, Guthridge JM, Brown EJ, Harley JB, James JA. Aberrant Epstein-Barr viral infection in systemic lupus erythematosus. Autoimmun Rev. 2009;8(4):337–42.
Sears J, Ujihara M, Wong S, Ott C, Middeldorp J, Aiyar A. The amino terminus of Epstein-Barr virus (EBV) nuclear antigen 1 contains AT hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes. J Virol. 2004;78(21):11487–505.
Dower SK. Cytokines, virokines and the evolution of immunity. Nat Immunol. 2000;1(5):367–8.
Mock A, Warta R, Geisenberger C, Bischoff R, Schulte A, Lamszus K, et al. Printed peptide arrays identify prognostic TNC serumantibodies in glioblastoma patients. Oncotarget. 2015;6(15):13579–90.
Maylin S, Feghoul L, Salmona M, Herda A, Mercier-Delarue S, Simon F, et al. Evaluation the architect EBV VCA IgM, VCA IgG, and EBNA-1 IgG chemiluminescent immunoassays to assess EBV serostatus prior transplantation. J Med Virol. 2017;89(11):2003–10.
Farina A, Cirone M, York M, Lenna S, Padilla C, McLaughlin S, et al. Epstein-Barr virus infection induces aberrant TLR activation pathway and fibroblast-myofibroblast conversion in scleroderma. J Invest Dermatol. 2014;134(4):954–64.
Farina A, Peruzzi G, Lacconi V, Lenna S, Quarta S, Rosato E, et al. Epstein-Barr virus lytic infection promotes activation of toll-like receptor 8 innate immune response in systemic sclerosis monocytes. Arthritis Res Ther. 2017;19(1):39.
Dreyfus DH, Liu Y, Ghoda LY, Chang JT. Analysis of an ankyrin-like region in Epstein Barr virus encoded (EBV) BZLF-1 (ZEBRA) protein: implications for interactions with NF-kappaB and p53. Virol J. 2011;8:422.
Dreyfus DH, Nagasawa M, Gelfand EW, Ghoda LY. Modulation of p53 activity by IkappaBalpha: evidence suggesting a common phylogeny between NF-kappaB and p53 transcription factors. BMC Immunol. 2005;6:12.
Herkel J, Mimran A, Erez N, Kam N, Lohse AW, Marker-Hermann E, et al. Autoimmunity to the p53 protein is a feature of systemic lupus erythematosus (SLE) related to anti-DNA antibodies. J Autoimmun. 2001;17(1):63–9.
Herkel J, Modrow JP, Bamberger S, Kanzler S, Rotter V, Cohen IR, et al. Prevalence of autoantibodies to the p53 protein in autoimmune hepatitis. Autoimmunity. 2002;35(8):493–6.
Leskowitz RM, Zhou XY, Villinger F, Fogg MH, Kaur A, Lieberman PM, et al. CD4+ and CD8+ T-cell responses to latent antigen EBNA-1 and lytic antigen BZLF-1 during persistent lymphocryptovirus infection of rhesus macaques. J Virol. 2013;87(15):8351–62.
Rubicz R, Yolken R, Drigalenko E, Carless MA, Dyer TD, Bauman L, et al. A genome-wide integrative genomic study localizes genetic factors influencing antibodies against Epstein-Barr virus nuclear antigen 1 (EBNA-1). PLoS Genet. 2013;9(1):e1003147.
Sheng Y, Saridakis V, Sarkari F, Duan S, Wu T, Arrowsmith CH, et al. Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat Struct Mol Biol. 2006;13(3):285–91.
Spanopoulou E, Zaitseva F, Wang FH, Santagata S, Baltimore D, Panayotou G. The homeodomain region of rag-1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell. 1996;87(2):263–76.
Yin FF, Bailey S, Innis CA, Ciubotaru M, Kamtekar S, Steitz TA, et al. Structure of the RAG1 nonamer binding domain with DNA reveals a dimer that mediates DNA synapsis. Nat Struct Mol Biol. 2009;16(5):499–508.
Dreyfus DH. Paleo-immunology: evidence consistent with insertion of a primordial herpes virus-like element in the origins of acquired immunity. PLoS One. 2009;4(6):e5778.
Hoebe EK, Hutajulu SH, van Beek J, Stevens SJ, Paramita DK, Greijer AE, et al. Purified hexameric Epstein-Barr virus-encoded BARF1 protein for measuring anti-BARF1 antibody responses in nasopharyngeal carcinoma patients. Clin Vaccine Immunol. 2011;18(2):298–304.
Kis LL, Takahara M, Nagy N, Klein G, Klein E. IL-10 can induce the expression of EBV-encoded latent membrane protein-1 (LMP-1) in the absence of EBNA-2 in B lymphocytes and in Burkitt lymphoma- and NK lymphoma-derived cell lines. Blood. 2006;107(7):2928–35.
Ouyang P, Rakus K, van Beurden SJ, Westphal AH, Davison AJ, Gatherer D, et al. IL-10 encoded by viruses: a remarkable example of independent acquisition of a cellular gene by viruses and its subsequent evolution in the viral genome. J Gen Virol. 2014;95(Pt 2):245–62.
Segal Y, Calabro M, Kanduc D, Shoenfeld Y. Human papilloma virus and lupus: the virus, the vaccine and the disease. Curr Opin Rheumatol. 2017;29(4):331–42.
Segal Y, Dahan S, Calabro M, Kanduc D, Shoenfeld Y. HPV and systemic lupus erythematosus: a mosaic of potential crossreactions. Immunol Res. 2017;65(2):564–71.
Segal Y, Dahan S, Sharif K, Bragazzi NL, Watad A, Amital H. The value of autoimmune syndrome induced by adjuvant (ASIA) - shedding light on orphan diseases in autoimmunity. Autoimmun Rev. 2018;17(5):440–8.
Segal Y, Shoenfeld Y. Vaccine-induced autoimmunity: the role of molecular mimicry and immune crossreaction. Cell Mol Immunol. 2018;15:586–94.
Segal Y, Zohar D, Shoenfeld Y. Infections and autoimmunity -new insights into an age-old reciprocity. Curr Opin Rheumatol. 2018;30(4):347–9.
Acknowledgements
The author acknowledges Yehuda Shoenfeld, MD and his research group (Tel Shomer, Sheba Medical Center, Israel, and Erwin Gelfand (retired), National Jewish Medical And Research Center, Denver CO) for their contributions to mentoring of DHD and helpful comments. Also, the authors acknowledge Eugenia Spanopoulou (deceased) for her unique and enduring contributions to the study of RAG protein A/T hooks and RAG function and Jaap Middeldorp for his pioneering work on EBV peptide arrays and helpful comment to the author of this work. Data processing and IgG binding analysis shown were provided on a complementary basis to Dr. David Dreyfus, Keren LLC, New Haven, CT by PEPperprint.com. All other analysis of the data was self-funded by the authors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
No conflict of interest reported by the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Dreyfus, D.H., Farina, A. & Farina, G.A. Molecular mimicry, genetic homology, and gene sharing proteomic “molecular fingerprints” using an EBV (Epstein-Barr virus)-derived microarray as a potential diagnostic method in autoimmune disease. Immunol Res 66, 686–695 (2018). https://doi.org/10.1007/s12026-018-9045-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12026-018-9045-0