Abstract
Lipid-specific T cells are important participants in human immune responses. Recognition of lipid antigens contributes to host defense against pathogens that can cause debilitating diseases, including mycobacterial, viral, and parasitic infections. Lipid-specific T cells also play important roles in various autoimmune diseases, atherosclerosis, and in tumor surveillance. A better understanding of the mechanisms that regulate lipid-reactive T-cell functions will enable the development of novel therapies across a wide range of diseases. In recent years, our laboratory has investigated lipid antigen specificities, mechanisms of lipid antigen presentation, molecular interaction of lipid antigens with CD1 antigen-presenting molecules, and the pathogenic and regulatory functions of lipid-specific T cells in a variety of disease settings. In this review, we present recent data that illustrate the critical role played by lipid-specific immune responses in host protection, with a particular focus on human studies.


Similar content being viewed by others
Abbreviations
- α-GalCer:
-
α-Galactosylceramide
- APC:
-
Antigen-presenting cells
- CDR:
-
Complementarity-determining region
- DC:
-
Dendritic cells
- ER:
-
Endoplasmic reticulum
- GSL:
-
Glycosphingolipid
- iNKT:
-
Semi-invariant natural killer T
- LPS:
-
Lipopolysaccharide
- LTP:
-
Lipid transfer protein
- PC:
-
Phosphatidylcholine
- TCR:
-
T-cell antigen receptor
- TGN:
-
Trans-Golgi network
- TLR:
-
Toll-like receptor
References
Porcelli S, Brenner MB, Greenstein JL, Balk SP, Terhorst C, Bleicher PA. Recognition of cluster of differentiation 1 antigens by human CD4−CD8− cytolytic T lymphocytes. Nature. 1989;341(6241):447–50.
Beckman EM, Porcelli SA, Morita CT, Behar SM, Furlong ST, Brenner MB. Recognition of a lipid antigen by CD1-restricted alpha beta+ T cells. Nature. 1994;372(6507):691–4.
Zeng Z, Castano AR, Segelke BW, Stura EA, Peterson PA, Wilson IA. Crystal structure of mouse CD1: an MHC-like fold with a large hydrophobic binding groove. Science. 1997;277(5324):339–45.
Dascher CC, Brenner MB. Evolutionary constraints on CD1 structure: insights from comparative genomic analysis. Trends Immunol. 2003;24(8):412–8.
Tourne S, Maitre B, Collmann A, Layre E, Mariotti S, Signorino-Gelo F, Loch C, Salamero J, Gilleron M, Angenieux C, Cazenave JP, Mori L, Hanau D, Puzo G, De Libero G, de la Salle H. Cutting edge: a naturally occurring mutation in CD1e impairs lipid antigen presentation. J Immunol. 2008;180(6):3642–6.
Porcelli S, Yockey CE, Brenner MB, Balk SP. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4−8− alpha/beta T cells demonstrates preferential use of several V beta genes and an invariant TCR alpha chain. J Exp Med. 1993;178(1):1–16.
Dellabona P, Padovan E, Casorati G, Brockhaus M, Lanzavecchia A. An invariant V alpha 24-J alpha Q/V beta 11 T cell receptor is expressed in all individuals by clonally expanded CD4−8− T cells. J Exp Med. 1994;180(3):1171–6.
Lantz O, Bendelac A. An invariant T cell receptor alpha chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4−8− T cells in mice and humans. J Exp Med. 1994;180(3):1097–106.
Godfrey DI, MacDonald HR, Kronenberg M, Smyth MJ, Van Kaer L. NKT cells: what’s in a name? Nat Rev Immunol. 2004;4(3):231–7.
De Libero G, Mori L. Recognition of lipid antigens by T cells. Nat Rev Immunol. 2005;5(6):485–96.
Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336.
Uldrich AP, Patel O, Cameron G, Pellicci DG, Day EB, Sullivan LC, Kyparissoudis K, Kjer-Nielsen L, Vivian JP, Cao B, Brooks AG, Williams SJ, Illarionov P, Besra GS, Turner SJ, Porcelli SA, McCluskey J, Smyth MJ, Rossjohn J, Godfrey DI. A semi-invariant Valpha10+ T cell antigen receptor defines a population of natural killer T cells with distinct glycolipid antigen-recognition properties. Nat Immunol. 2011;12(7):616–23.
Cardell S, Tangri S, Chan S, Kronenberg M, Benoist C, Mathis D. CD1-restricted CD4+ T cells in major histocompatibility complex class II-deficient mice. J Exp Med. 1995;182(4):993–1004.
Exley MA, He Q, Cheng O, Wang RJ, Cheney CP, Balk SP, Koziel MJ. Cutting edge: compartmentalization of Th1-like noninvariant CD1d-reactive T cells in hepatitis C virus-infected liver. J Immunol. 2002;168(4):1519–23.
Moody DB, Zajonc DM, Wilson IA. Anatomy of CD1-lipid antigen complexes. Nat Rev Immunol. 2005;5(5):387–99.
De Libero G, Mori L. The easy virtue of CD1c. Immunity. 2010;33(6):831–3.
Zajonc DM, Elsliger MA, Teyton L, Wilson IA. Crystal structure of CD1a in complex with a sulfatide self antigen at a resolution of 2.15 A. Nat Immunol. 2003;4(8):808–15.
Shamshiev A, Gober HJ, Donda A, Mazorra Z, Mori L, De Libero G. Presentation of the same glycolipid by different CD1 molecules. J Exp Med. 2002;195(8):1013–21.
Manolova V, Kistowska M, Paoletti S, Baltariu GM, Bausinger H, Hanau D, Mori L, De Libero G. Functional CD1a is stabilized by exogenous lipids. Eur J Immunol. 2006;36(5):1083–92.
Gadola SD, Zaccai NR, Harlos K, Shepherd D, Castro-Palomino JC, Ritter G, Schmidt RR, Jones EY, Cerundolo V. Structure of human CD1b with bound ligands at 2.3 A, a maze for alkyl chains. Nat Immunol. 2002;3(8):721–6.
Cheng TY, Relloso M, Van Rhijn I, Young DC, Besra GS, Briken V, Zajonc DM, Wilson IA, Porcelli S, Moody DB. Role of lipid trimming and CD1 groove size in cellular antigen presentation. Embo J. 2006;25(13):2989–99.
Ernst WA, Maher J, Cho S, Niazi KR, Chatterjee D, Moody DB, Besra GS, Watanabe Y, Jensen PE, Porcelli SA, Kronenberg M, Modlin RL. Molecular interaction of CD1b with lipoglycan antigens. Immunity. 1998;8(3):331–40.
Kronenberg M, Sullivan BA. Acid test: lipid antigens get into the groove. Immunity. 2008;28(6):727–9.
Garcia-Alles LF, Versluis K, Maveyraud L, Vallina AT, Sansano S, Bello NF, Gober HJ, Guillet V, de la Salle H, Puzo G, Mori L, Heck AJ, De Libero G, Mourey L. Endogenous phosphatidylcholine and a long spacer ligand stabilize the lipid-binding groove of CD1b. EMBO J. 2006;25(15):3684–92.
Garcia-Alles LF, Collmann A, Versluis C, Lindner B, Guiard J, Maveyraud L, Huc E, Im JS, Sansano S, Brando T, Julien S, Prandi J, Gilleron M, Porcelli SA, de la Salle H, Heck AJ, Mori L, Puzo G, Mourey L, De Libero G. Structural reorganization of the antigen-binding groove of human CD1b for presentation of mycobacterial sulfoglycolipids. Proc Natl Acad Sci USA. 2011;108(43):17755–60.
Scharf L, Li NS, Hawk AJ, Garzon D, Zhang T, Fox LM, Kazen AR, Shah S, Haddadian EJ, Gumperz JE, Saghatelian A, Faraldo-Gomez JD, Meredith SC, Piccirilli JA, Adams EJ. The 2.5 a structure of CD1c in complex with a mycobacterial lipid reveals an open groove ideally suited for diverse antigen presentation. Immunity. 2010;33(6):853–62.
Koch M, Stronge VS, Shepherd D, Gadola SD, Mathew B, Ritter G, Fersht AR, Besra GS, Schmidt RR, Jones EY, Cerundolo V. The crystal structure of human CD1d with and without alpha-galactosylceramide. Nat Immunol. 2005;6(8):819–26.
Zajonc DM, Wilson IA. Architecture of CD1 proteins. Curr Top Microbiol Immunol. 2007;314:27–50.
Angenieux C, Fraisier V, Maitre B, Racine V, van der Wel N, Fricker D, Proamer F, Sachse M, Cazenave JP, Peters P, Goud B, Hanau D, Sibarita JB, Salamero J, de la Salle H. The cellular pathway of CD1e in immature and maturing dendritic cells. Traffic. 2005;6(4):286–302.
Garcia-Alles LF, Giacometti G, Versluis C, Maveyraud L, de Paepe D, Guiard J, Tranier S, Gilleron M, Prandi J, Hanau D, Heck AJ, Mori L, De Libero G, Puzo G, Mourey L, de la Salle H. Crystal structure of human CD1e reveals a groove suited for lipid-exchange processes. Proc Natl Acad Sci USA. 2011;108(32):13230–5.
de la Salle H, Mariotti S, Angenieux C, Gilleron M, Garcia-Alles LF, Malm D, Berg T, Paoletti S, Maitre B, Mourey L, Salamero J, Cazenave JP, Hanau D, Mori L, Puzo G, De Libero G. Assistance of microbial glycolipid antigen processing by CD1e. Science. 2005;310(5752):1321–4.
Facciotti F, Cavallari M, Angenieux C, Garcia-Alles LF, Signorino-Gelo F, Angman L, Gilleron M, Prandi J, Puzo G, Panza L, Xia C, Wang PG, Dellabona P, Casorati G, Porcelli SA, de la Salle H, Mori L, De Libero G. Fine tuning by human CD1e of lipid-specific immune responses. Proc Natl Acad Sci USA. 2011;108(34):14228–33.
Borg NA, Wun KS, Kjer-Nielsen L, Wilce MC, Pellicci DG, Koh R, Besra GS, Bharadwaj M, Godfrey DI, McCluskey J, Rossjohn J. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 2007;448(7149):44–9.
Mallevaey T, Scott-Browne JP, Matsuda JL, Young MH, Pellicci DG, Patel O, Thakur M, Kjer-Nielsen L, Richardson SK, Cerundolo V, Howell AR, McCluskey J, Godfrey DI, Rossjohn J, Marrack P, Gapin L. T cell receptor CDR2 beta and CDR3 beta loops collaborate functionally to shape the iNKT cell repertoire. Immunity. 2009;31(1):60–71.
Wun KS, Cameron G, Patel O, Pang SS, Pellicci DG, Sullivan LC, Keshipeddy S, Young MH, Uldrich AP, Thakur MS, Richardson SK, Howell AR, Illarionov PA, Brooks AG, Besra GS, McCluskey J, Gapin L, Porcelli SA, Godfrey DI, Rossjohn J. A molecular basis for the exquisite CD1d-restricted antigen specificity and functional responses of natural killer T cells. Immunity. 2011;34(3):327–39.
Cantu C III, Benlagha K, Savage PB, Bendelac A, Teyton L. The paradox of immune molecular recognition of alpha-galactosylceramide: low affinity, low specificity for CD1d, high affinity for alpha beta TCRs. J Immunol. 2003;170(9):4673–82.
Sidobre S, Hammond KJ, Benazet-Sidobre L, Maltsev SD, Richardson SK, Ndonye RM, Howell AR, Sakai T, Besra GS, Porcelli SA, Kronenberg M. The T cell antigen receptor expressed by Valpha14i NKT cells has a unique mode of glycosphingolipid antigen recognition. Proc Natl Acad Sci USA. 2004;101(33):12254–9.
Gadola SD, Koch M, Marles-Wright J, Lissin NM, Shepherd D, Matulis G, Harlos K, Villiger PM, Stuart DI, Jakobsen BK, Cerundolo V, Jones EY. Structure and binding kinetics of three different human CD1d-alpha-galactosylceramide-specific T cell receptors. J Exp Med. 2006;203(3):699–710.
McCarthy C, Shepherd D, Fleire S, Stronge VS, Koch M, Illarionov PA, Bossi G, Salio M, Denkberg G, Reddington F, Tarlton A, Reddy BG, Schmidt RR, Reiter Y, Griffiths GM, van der Merwe PA, Besra GS, Jones EY, Batista FD, Cerundolo V. The length of lipids bound to human CD1d molecules modulates the affinity of NKT cell TCR and the threshold of NKT cell activation. J Exp Med. 2007;204(5):1131–44.
Aleksic M, Dushek O, Zhang H, Shenderov E, Chen JL, Cerundolo V, Coombs D, van der Merwe PA. Dependence of T cell antigen recognition on T cell receptor-peptide MHC confinement time. Immunity. 2010;32(2):163–74.
Stanic AK, Shashidharamurthy R, Bezbradica JS, Matsuki N, Yoshimura Y, Miyake S, Choi EY, Schell TD, Van Kaer L, Tevethia SS, Roopenian DC, Yamamura T, Joyce S. Another view of T cell antigen recognition: cooperative engagement of glycolipid antigens by Va14Ja18 natural T(iNKT) cell receptor [corrected]. J Immunol. 2003;171(9):4539–51.
Pellicci DG, Patel O, Kjer-Nielsen L, Pang SS, Sullivan LC, Kyparissoudis K, Brooks AG, Reid HH, Gras S, Lucet IS, Koh R, Smyth MJ, Mallevaey T, Matsuda JL, Gapin L, McCluskey J, Godfrey DI, Rossjohn J. Differential recognition of CD1d-alpha-galactosyl ceramide by the V beta 8.2 and V beta 7 semi-invariant NKT T cell receptors. Immunity. 2009;31(1):47–59.
Brennan PJ, Tatituri RV, Brigl M, Kim EY, Tuli A, Sanderson JP, Gadola SD, Hsu FF, Besra GS, Brenner MB. Invariant natural killer T cells recognize lipid self antigen induced by microbial danger signals. Nat Immunol. 2011;12(12):1202–11.
Li Y, Girardi E, Wang J, Yu ED, Painter GF, Kronenberg M, Zajonc DM. The Valpha14 invariant natural killer T cell TCR forces microbial glycolipids and CD1d into a conserved binding mode. J Exp Med. 2010;207(11):2383–93.
Melian A, Watts GF, Shamshiev A, De Libero G, Clatworthy A, Vincent M, Brenner MB, Behar S, Niazi K, Modlin RL, Almo S, Ostrov D, Nathenson SG, Porcelli SA. Molecular recognition of human CD1b antigen complexes: evidence for a common pattern of interaction with alpha beta TCRs. J Immunol. 2000;165(8):4494–504.
Barral DC, Brenner MB. CD1 antigen presentation: how it works? Nat Rev Immunol. 2007;7(12):929–41.
Thomas SY, Hou R, Boyson JE, Means TK, Hess C, Olson DP, Strominger JL, Brenner MB, Gumperz JE, Wilson SB, Luster AD. CD1d-restricted NKT cells express a chemokine receptor profile indicative of Th1-type inflammatory homing cells. J Immunol. 2003;171(5):2571–80.
Wang X, Chen X, Rodenkirch L, Simonson W, Wernimont S, Ndonye RM, Veerapen N, Gibson D, Howell AR, Besra GS, Painter GF, Huttenlocher A, Gumperz JE. Natural killer T-cell autoreactivity leads to a specialized activation state. Blood. 2008;112(10):4128–38.
Stetson DB, Mohrs M, Reinhardt RL, Baron JL, Wang ZE, Gapin L, Kronenberg M, Locksley RM. Constitutive cytokine mRNAs mark natural killer (NK) and NK T cells poised for rapid effector function. J Exp Med. 2003;198(7):1069–76.
Brigl M, Tatituri RV, Watts GF, Bhowruth V, Leadbetter EA, Barton N, Cohen NR, Hsu FF, Besra GS, Brenner MB. Innate and cytokine-driven signals, rather than microbial antigens, dominate in natural killer T cell activation during microbial infection. J Exp Med. 2011;208(6):1163–77.
De Libero G, Moran AP, Gober HJ, Rossy E, Shamshiev A, Chelnokova O, Mazorra Z, Vendetti S, Sacchi A, Prendergast MM, Sansano S, Tonevitsky A, Landmann R, Mori L. Bacterial infections promote T cell recognition of self-glycolipids. Immunity. 2005;22(6):763–72.
Nagarajan NA, Kronenberg M. Invariant NKT cells amplify the innate immune response to lipopolysaccharide. J Immunol. 2007;178(5):2706–13.
Tyznik AJ, Tupin E, Nagarajan NA, Her MJ, Benedict CA, Kronenberg M. Cutting edge: the mechanism of invariant NKT cell responses to viral danger signals. J Immunol. 2008;181(7):4452–6.
Wesley JD, Tessmer MS, Chaukos D, Brossay L. NK cell-like behavior of Valpha14i NK T cells during MCMV infection. PLoS Pathog. 2008;4(7):e1000106.
de Jong A, Pena-Cruz V, Cheng TY, Clark RA, Van Rhijn I, Moody DB. CD1a-autoreactive T cells are a normal component of the human alphabeta T cell repertoire. Nat Immunol. 2010;11(12):1102–9.
de Lalla C, Lepore M, Piccolo FM, Rinaldi A, Scelfo A, Garavaglia C, Mori L, De Libero G, Dellabona P, Casorati G. High-frequency and adaptive-like dynamics of human CD1 self-reactive T cells. Eur J Immunol. 2011;41(3):602–10.
Vincent MS, Xiong X, Grant EP, Peng W, Brenner MB. CD1a-, b-, and c-restricted TCRs recognize both self and foreign antigens. J Immunol. 2005;175(10):6344–51.
Blomqvist M, Rhost S, Teneberg S, Lofbom L, Osterbye T, Brigl M, Mansson JE, Cardell SL. Multiple tissue-specific isoforms of sulfatide activate CD1d-restricted type II NKT cells. Eur J Immunol. 2009;39(7):1726–35.
Shamshiev A, Donda A, Carena I, Mori L, Kappos L, De Libero G. Self glycolipids as T-cell autoantigens. Eur J Immunol. 1999;29(5):1667–75.
Fox LM, Cox DG, Lockridge JL, Wang X, Chen X, Scharf L, Trott DL, Ndonye RM, Veerapen N, Besra GS, Howell AR, Cook ME, Adams EJ, Hildebrand WH, Gumperz JE. Recognition of lyso-phospholipids by human natural killer T lymphocytes. PLoS Biol. 2009;7(10):e1000228.
Shamshiev A, Donda A, Prigozy TI, Mori L, Chigorno V, Benedict CA, Kappos L, Sonnino S, Kronenberg M, De Libero G. The alphabeta T cell response to self-glycolipids shows a novel mechanism of CD1b loading and a requirement for complex oligosaccharides. Immunity. 2000;13(2):255–64.
Wu DY, Segal NH, Sidobre S, Kronenberg M, Chapman PB. Cross-presentation of disialoganglioside GD3 to natural killer T cells. J Exp Med. 2003;198(1):173–81.
Zhou D, Mattner J, Cantu C III, Schrantz N, Yin N, Gao Y, Sagiv Y, Hudspeth K, Wu YP, Yamashita T, Teneberg S, Wang D, Proia RL, Levery SB, Savage PB, Teyton L, Bendelac A. Lysosomal glycosphingolipid recognition by NKT cells. Science. 2004;306(5702):1786–9.
Dieude M, Striegl H, Tyznik AJ, Wang J, Behar SM, Piccirillo CA, Levine JS, Zajonc DM, Rauch J. Cardiolipin binds to CD1d and stimulates CD1d-restricted gammadelta T cells in the normal murine repertoire. J Immunol. 2011;186(8):4771–81.
Gumperz JE, Roy C, Makowska A, Lum D, Sugita M, Podrebarac T, Koezuka Y, Porcelli SA, Cardell S, Brenner MB, Behar SM. Murine CD1d-restricted T cell recognition of cellular lipids. Immunity. 2000;12(2):211–21.
Rauch J, Gumperz J, Robinson C, Skold M, Roy C, Young DC, Lafleur M, Moody DB, Brenner MB, Costello CE, Behar SM. Structural features of the acyl chain determine self-phospholipid antigen recognition by a CD1d-restricted invariant NKT (iNKT) cell. J Biol Chem. 2003;278(48):47508–15.
Agea E, Russano A, Bistoni O, Mannucci R, Nicoletti I, Corazzi L, Postle AD, De Libero G, Porcelli SA, Spinozzi F. Human CD1-restricted T cell recognition of lipids from pollens. J Exp Med. 2005;202(2):295–308.
Facciotti F, Ramanjaneyulu GS, Lepore M, Sansano S, Ni G, Forss-Petter S, Berger J, Xia C, Mori L, De libero G. Peroxisome-derived lipids are self-antigens stimulating invariant Natural Killer T cells in the thymus. Nat Immunol. 2012; in press.
Gilleron M, Stenger S, Mazorra Z, Wittke F, Mariotti S, Bohmer G, Prandi J, Mori L, Puzo G, De Libero G. Diacylated sulfoglycolipids are novel mycobacterial antigens stimulating CD1-restricted T cells during infection with Mycobacterium tuberculosis. J Exp Med. 2004;199(5):649–59.
Sieling PA, Chatterjee D, Porcelli SA, Prigozy TI, Mazzaccaro RJ, Soriano T, Bloom BR, Brenner MB, Kronenberg M, Brennan PJ, et al. CD1-restricted T cell recognition of microbial lipoglycan antigens. Science. 1995;269(5221):227–30.
Layre E, Collmann A, Bastian M, Mariotti S, Czaplicki J, Prandi J, Mori L, Stenger S, De Libero G, Puzo G, Gilleron M. Mycolic acids constitute a scaffold for mycobacterial lipid antigens stimulating CD1-restricted T cells. Chem Biol. 2009;16(1):82–92.
Moody DB, Ulrichs T, Muhlecker W, Young DC, Gurcha SS, Grant E, Rosat JP, Brenner MB, Costello CE, Besra GS, Porcelli SA. CD1c-mediated T-cell recognition of isoprenoid glycolipids in Mycobacterium tuberculosis infection. Nature. 2000;404(6780):884–8.
Matsunaga I, Bhatt A, Young DC, Cheng TY, Eyles SJ, Besra GS, Briken V, Porcelli SA, Costello CE, Jacobs WR Jr, Moody DB. Mycobacterium tuberculosis pks12 produces a novel polyketide presented by CD1c to T cells. J Exp Med. 2004;200(12):1559–69.
Moody DB, Young DC, Cheng TY, Rosat JP, Roura-Mir C, O’Connor PB, Zajonc DM, Walz A, Miller MJ, Levery SB, Wilson IA, Costello CE, Brenner MB. T cell activation by lipopeptide antigens. Science. 2004;303(5657):527–31.
Kinjo Y, Tupin E, Wu D, Fujio M, Garcia-Navarro R, Benhnia MR, Zajonc DM, Ben-Menachem G, Ainge GD, Painter GF, Khurana A, Hoebe K, Behar SM, Beutler B, Wilson IA, Tsuji M, Sellati TJ, Wong CH, Kronenberg M. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat Immunol. 2006;7(9):978–86.
Kinjo Y, Illarionov P, Vela JL, Pei B, Girardi E, Li X, Li Y, Imamura M, Kaneko Y, Okawara A, Miyazaki Y, Gomez-Velasco A, Rogers P, Dahesh S, Uchiyama S, Khurana A, Kawahara K, Yesilkaya H, Andrew PW, Wong CH, Kawakami K, Nizet V, Besra GS, Tsuji M, Zajonc DM, Kronenberg M. Invariant natural killer T cells recognize glycolipids from pathogenic gram-positive bacteria. Nat Immunol. 2011;12(10):966–74.
Chang YJ, Kim HY, Albacker LA, Lee HH, Baumgarth N, Akira S, Savage PB, Endo S, Yamamura T, Maaskant J, Kitano N, Singh A, Bhatt A, Besra GS, van den Elzen P, Appelmelk B, Franck RW, Chen G, DeKruyff RH, Shimamura M, Illarionov P, Umetsu DT. Influenza infection in suckling mice expands an NKT cell subset that protects against airway hyperreactivity. J Clin Invest. 2011;121(1):57–69.
Mattner J, DeBord KL, Ismail N, Goff RD, Cantu C III, Zhou D, Saint-Mezard P, Wang V, Gao Y, Yin N, Hoebe K, Schneewind O, Walker D, Beutler B, Teyton L, Savage PB, Bendelac A. Both exogenous and endogenous glycolipid antigens activate N KT cells during microbial infections. Nature. 2005;434:525–9.
Kinjo Y, Wu D, Kim G, Xing G-W, Poles MA, Ho DH, Tsuji M, Kawahara K, Wong C-H, Kronenberg M. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature. 2005;434:520–5.
Sriram V, Du W, Gervay-Hague J, Brutkiewicz RR. Cell wall glycosphingolipids of Sphingomonas paucimobilis are CD1d-specific ligands for NKT cells. Eur J Immunol. 2005;35(6):1692–701.
Mattner J, Savage PB, Leung P, Oertelt SS, Wang V, Trivedi O, Scanlon ST, Pendem K, Teyton L, Hart J, Ridgway WM, Wicker LS, Gershwin ME, Bendelac A. Liver autoimmunity triggered by microbial activation of natural killer T cells. Cell Host Microbe. 2008;3(5):304–15.
Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, Ueno H, Nakagawa R, Sato H, Kondo E, Koseki H, Taniguchi M. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 1997;278(5343):1626–9.
Van Rhijn I, Young DC, Im JS, Levery SB, Illarionov PA, Besra GS, Porcelli SA, Gumperz J, Cheng TY, Moody DB. CD1d-restricted T cell activation by nonlipidic small molecules. Proc Natl Acad Sci USA. 2004;101(37):13578–83.
Acknowledgments
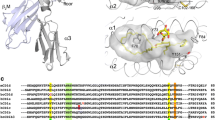
We thank Anil Kumar for the graphic elaboration of the structures.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mori, L., De Libero, G. T cells specific for lipid antigens. Immunol Res 53, 191–199 (2012). https://doi.org/10.1007/s12026-012-8294-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12026-012-8294-6