Abstract
Purpose
The length of time a critically ill coronavirus disease 2019 (COVID-19) patient remains infectious and should therefore be isolated remains unknown. This prospective study was undertaken in critically ill patients to evaluate the reliability of single negative real-time polymerase chain reaction (RT-PCR) in lower tracheal aspirates (LTA) in predicting a second negative test and to analyze clinical factors potentially influencing the viral shedding.
Methods
From April 9, 2020 onwards, intubated COVID-19 patients treated in the intensive care unit were systematically evaluated for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by RT-PCR of nasopharyngeal swabs and LTA. The time to negativity was defined as the time between the onset of symptoms and the viral clearance in LTA. In order to identify risk factors for prolonged viral shedding, we used univariate and multivariate Cox proportional hazards models.
Results
Forty-eight intubated SARS-CoV-2 patients were enrolled. Overall, we observed that the association of the first negative RT-PCR with a second negative result was 96.7%. Median viral shedding was 25 (IQR: 21.5–28) days since symptoms’ onset. In the univariate Cox model analysis, type 2 diabetes mellitus was associated with a prolonged viral RNA shedding (hazard ratio [HR]: 0.41, 95% CI: 0.06–3.11, p = 0.04). In the multivariate Cox model analysis, type 2 diabetes was associated with a prolonged viral RNA shedding (HR: 0.31, 95% CI: 0.11–0.89, p = 0.029).
Conclusion
Intubated patients with type 2 diabetes mellitus may have prolonged SARS-CoV-2 shedding. In critically ill COVID-19 patients, one negative LTA should be sufficient to assess and exclude infectivity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Treatment of critically ill coronavirus disease 2019 (COVID-19) patients requires strict isolation to limit nosocomial spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). However, to date, the length of time an individual infected with SARS-CoV-2 remains infectious and needs to be isolated remains unknown, as viral shedding has been described to persist for an interval varying between 10 [1] and 60 days [2] after symptoms’ onset. Mild cases may have an earlier viral clearance (i.e., negative real-time polymerase chain reaction (RT-PCR) or cell cultures by day 10 post onset) [1, 3], whereas severe cases may have longer viral shedding [4, 5]. Most studies focused on nasopharyngeal or oropharyngeal swabs [3, 4, 6,7,8,9] and did not consider lower respiratory samples. Most importantly in this context, only one study (until June 2020) assessed the infectivity with cell culture for SARS-CoV-2 [1].
This prospective study was undertaken in critically ill patients to (1) evaluate the reliability of single negative RT-PCR in predicting a second negative test, (2) investigate the interval from symptoms’ onset to RT-PCR negativity, and (3) analyze clinical factors potentially influencing the latter.
Methods
Geographic and demographic context
The study was performed at Locarno community hospital, a 250 beds facility entirely dedicated to COVID-19 patients during the SARS-CoV-2 pandemic, and is part of the public hospital network of southern Switzerland, serving an area of 350,000 inhabitants and being close to the SARS-CoV-2 outbreak epicenter of Northern Italy (Fig. 1). All data of SARS-CoV-2 patients were collected in a specific standardized institutional database.
Institutional management of intubated SARS-Cov-2 patients
From April 9, 2020 onwards, COVID-19 patients treated in the intensive care unit (ICU) were systematically evaluated for SARS-CoV-2 by RT-PCR of nasopharyngeal swabs (NPS) and lower tracheal aspirates (LTA). After their initial test, all patients underwent a biweekly LTA RT-PCR reevaluation until removal of the endotracheal tube or death or, according to the literature [10], until two consecutive negative RT-PCR results (Fig. 2). Furthermore, in selected cases and in order to prove the true negativity of a single negative RT-PCR, a viral culture was performed. In the included patients the results of NPS were also routinely collected.
Timeline of RT-PCR evaluation according to the institutional management during the SARS-CoV-2 pandemic. NPS nasopharyngeal sample, LTA lower tracheal aspirate. Point prevalence: LTA and NPS were performed at 2 consecutive days in all intubated patients. During the follow-up NPS were performed only if an NPS was positive during the point prevalence assessment
Patient selection and variables collected
All intubated ICU patients hospitalized at our institution between April 9 and May 12, 2020 were retrospectively reviewed. The following variables were routinely collected: demographic, clinical, laboratory, and microbiological data on SARS-CoV-2 patients, ICU-specific complications, and information on management. Mortality was assessed on May 20, 2020.
Microbiological analyses: RT-PCR and viral cultures
Sampling was performed according to a designed protocol. Swab samples were immediately inserted into sterile tubes containing 3 mL UTM-RT® viral transport medium (Copan, Brescia Italy), LTA were placed into sterile tubes. Both types of specimens were sent to the microbiology laboratory for sample processing and viral RNA extraction, which were performed on the same day. RT-PCR on NPS were performed using the commercial kit SARS-CoV-2 S Gene VIASURE Real Time PCR detection kit by CerTest BIOTEC on BD MAX Instrument (Becton Dickinson, New Jersey, USA). RNA extracted from LTA were amplified using a second PCR system based on the protocol published by Corman et al. [11] with primers and probes produced by TibMol Biol (Berlin, Germany). The viral load was indicated as cycle threshold (Ct) value of S gene of SARS-CoV-2 (VIASURE) and E- and RdRP gene, respectively. A positive and a negative control, as well as internal controls, was included in the assay, according to the manufacturer’s protocol. A Ct value of <40 was defined as positive for SARS-CoV-2 RNA and >40 was defined as negative. Samples with a Ct value between 37 and 40 were retested, at least twice.
For virus cultures, patient material was precleared by centrifugation and filtered before inoculation of Vero CCL81 cells with a dilution series of the patient material. Cells were observed daily for cytopathic effects (CPE) and upon detection of CPE or at latest by d7 supernatants were harvested and tested for the presence of SARS-CoV-2 by RT-qPCR.
Outcome
The onset of symptoms was used as the starting time point for the viral clearance process. The date of the first negative detection of viral RNA (i.e., time to negativity or viral shedding) was defined as the end time point of viral clearance (see “Results” section).
Statistical analyses
The time to negativity was described using descriptive statistics (median and IQR). In order to identify risk factors for prolonged viral shedding, we used univariate Cox proportional hazards models. To date, no study exhaustively investigated risk factors for prolonged viral shedding and, therefore, we performed multivariate Cox models including variables that showed p values < 0.20 in the univariate analysis. A hazard ratio (HR) of <1 indicated prolonged viral RNA shedding. In the Cox models, the proportionality of hazard risks was tested using Martingale residuals. The graphical of daily risk for prolonged viral shedding was illustrated using the hazard rate function from right-censored data using kernel-based methods [12]. All statistical analyses were performed with SAS (version 9.4).
Results
Demographic features
According to the study design and selection criteria, 48 intubated SARS-CoV-2 patients were enrolled. Overall, 161 RT-PCR tests of LTA were performed and patients underwent two to eight evaluations. The median time from onset of symptoms and the first sampling was 28 days (IQR: 23.5–32.5). Table 1 details the characteristics of the patients. Twelve patients (25%) patients died until May 20, 2020. At screening time, 31 patients (65%) showed same results between NPS and LTA, whereas in 15 patients (31%) LTA samples were positive and NPS were negative.
Evaluation of single negative RT-PCR as predictor of second negative test
In total, 31 patients with one negative RT-PCR in LTA during their follow-up and 28 (90.3%) had a second negative test within the following 3 days. Two of the remaining three cases had a negative test within 1 week. In one case, the subsequent RT-PCR was positive with 37-Ct. Among these 31 patients, data on the viral culture of LTA samples were available in four cases at their time of first negative RT-PCR and all these patients had both negative RT-PCR and culture results. Overall, we observed that the association of the first negative RT-PCR with a second negative result was 96.7%.
Analysis of viral shedding
Based on the above results, we are confident that a single negative RT-PCR in LTA can serve as a proof of negativity. Median viral shedding was 25 (IQR 21.5–28) days since symptoms’ onset (Table 1).
Risk factors for prolonged viral shedding
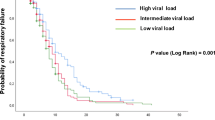
In the univariate Cox model analysis, type 2 diabetes mellitus was associated with a prolonged viral RNA shedding (HR: 0.41, 95% CI: 0.06–3.11, p = 0.04, Table 2 and Fig. 3).
On the contrary, lymphopenia duration (HR: 0.98, 95% CI: 0.94–1.01, p = 0.23), ventilator-associated pneumonia (VAP) during ICU stay (HR: 1.33, 95% CI: 0.38–1.66, p = 0.54), severe ARDS (HR: 0.59, 95% CI: 0.25–1.39, p = 0.23), and antivirals (HR: 0.63, 95% CI: 0.30–1.33, p = 0.23) did not significantly influence the duration of viral shedding. Also in the multivariate Cox model analysis type 2 diabetes mellitus was associated with a prolonged viral RNA shedding (HR: 0.31, 95% CI: 0.11–0.89, p = 0.029, Table 2), and similar results were observed in a sensitivity analysis defining negativity using two negative LTA samples (HR: 0.23, 95% CI: 0.07–0.69, p = 0.0089, Table 3). Of note, the mortality was higher among type 2 diabetes patients (54 versus 14%, p < 0.01).
Discussion
This study highlighted two important points concerning viral shedding in critically ill COVID-19 patients: (1) type 2 diabetes mellitus is associated with prolonged SARS-CoV-2 shedding; (2) a first negative LTA sample is predictive for a second negative sample.
Type 2 diabetes patients are at the highest risk for complications from COVID-19 infection and several authors described the relationship between COVID-19 and diabetes [13, 14]. We showed that type 2 diabetes mellitus was identified as a risk factor for prolonged SARS-CoV-2 shedding. We suggest the following possible pathophysiological explanations: (1) the innate immune system is the first line of defense against SARS-CoV-2, and may be compromised in patients with uncontrolled type 2 diabetes [15, 16]; (2) patients with type 2 diabetes mellitus may be more susceptible to an inflammatory cytokine storm eventually leading to rapid deterioration of COVID-19 [17] and to reduced control of viral shedding; (3) type 2 diabetes mellitus reduced the expression of angiotensin-converting enzyme 2 which may play a potent anti-inflammatory and antioxidant role in the lung and, probably, may prolong viral shedding [18]. Therefore, critically ill people with type 2 diabetes who are infected with COVID-19 may require a prolonged infection prevention measures during the hospitalization. Interestingly, corticosteroid therapy during ICU stay, VAP, antiviral therapy, lymphopenia, and severe ARDS did not significantly influence the duration of viral shedding in our study population.
The ECDC recommends two consecutive negative RT-PCR tests from respiratory specimens at 24 h interval at least 8 days after symptoms onset to discontinue isolation precautions in hospitalized COVID-19 patients [10]. Similar recommendations were issued by several institutions in other continents [5, 19]. Specimens were usually collected in the upper respiratory tract, and data on lower respiratory samples are rarely considered. Viral shedding from upper respiratory tract appeared to be higher soon after symptoms’ onset; however, during the course of disease, viral shedding is predominantly located in the lower respiratory tract [1]. Using lower respiratory tract samples, we showed that one negative RT-PCR LTA sample is sufficient to confirm the absence of viral shedding in critically ill intubated COVID-19 patients, and the results of RT-PCR were also confirmed in selected patients by viral cell cultures. These findings highlight the importance of virological monitoring of LTA samples in critically ill patients to decide when isolation precautions for the prevention of respiratory infections could be discontinued, thus making an early transfer to general wards possible, and encouraging an early rehabilitation program that may accelerate the discharge at home and the reintegration into society.
Our study has several limitations. First, we performed an observational study using surveillance data. Second, LTA samples were not collected on a daily basis. Third, viral cell cultures were not routinely performed during the study. It is therefore conceivable that the results on viral shedding might have been different if this method would have been used systematically. Third, due to the relatively small sample size, the statistical analyses should be interpreted with caution. Finally, we did not collect further LTA samples after two negative results.
Intubated patients with type 2 diabetes mellitus may have prolonged SARS-CoV-2 shedding. In critically ill COVID-19 patients, one negative LTA should be sufficient to assess and exclude infectivity.
Data availability
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
R. Wolfel, V.M. Corman, W. Guggemos, M. Seilmaier, S. Zange, M.A. Muller, D. Niemeyer, T.C. Jones, P. Vollmar, C. Rothe, M. Hoelscher, T. Bleicker, S. Brunink, J. Schneider, R. Ehmann, K. Zwirglmaier, C. Drosten, C. Wendtner, Virological assessment of hospitalized patients with COVID-2019. Nature (2020). https://doi.org/10.1038/s41586-020-2196-x
J. Li, L. Zhang, B. Liu, D. Song, Case report: viral shedding for 60 days in a woman with novel coronavirus disease (COVID-19). Am. J Trop. Med. Hyg. (2020). https://doi.org/10.4269/ajtmh.20-0275
Y. Liu, L.M. Yan, L. Wan, T.X. Xiang, A. Le, J.M. Liu, M. Peiris, L.L.M. Poon, W. Zhang, Viral dynamics in mild and severe cases of COVID-19. Lancet Infect. Dis. (2020). https://doi.org/10.1016/S1473-3099(20)30232-2
B. Zhou, J. She, Y. Wang, X. Ma, The duration of viral shedding of discharged patients with severe COVID-19. Clin. Infect. Dis. (2020). https://doi.org/10.1093/cid/ciaa451
Y. Huang, S. Chen, Z. Yang, W. Guan, D. Liu, Z. Lin, Y. Zhang, Z. Xu, X. Liu, Y. Li, SARS-CoV-2 viral load in clinical samples of critically Ill patients. Am. J. Respir. Crit. Care Med. (2020). https://doi.org/10.1164/rccm.202003-0572LE
A.T. Xiao, Y.X. Tong, S. Zhang, Profile of RT-PCR for SARS-CoV-2: a preliminary study from 56 COVID-19 patients. Clin. Infect. Dis. (2020). https://doi.org/10.1093/cid/ciaa460
S. Zheng, J. Fan, F. Yu, B. Feng, B. Lou, Q. Zou, G. Xie, S. Lin, R. Wang, X. Yang, W. Chen, Q. Wang, D. Zhang, Y. Liu, R. Gong, Z. Ma, S. Lu, Y. Xiao, Y. Gu, J. Zhang, H. Yao, K. Xu, X. Lu, G. Wei, J. Zhou, Q. Fang, H. Cai, Y. Qiu, J. Sheng, Y. Chen, T. Liang, Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ 369, m1443 (2020). https://doi.org/10.1136/bmj.m1443
D. Yan, X.Y. Liu, Y.N. Zhu, L. Huang, B.T. Dan, G.J. Zhang, Y.H. Gao, Factors associated with prolonged viral shedding and impact of Lopinavir/Ritonavir treatment in hospitalised non-critically ill patients with SARS-CoV-2 infection. Eur. Respir. J. (2020). https://doi.org/10.1183/13993003.00799-2020
L. Qi, Y. Yang, D. Jiang, C. Tu, L. Wan, X. Chen, Z. Li, Factors associated with duration of viral shedding in adults with COVID-19 outside of Wuhan, China: a retrospective cohort study. Int. J. Infect. Dis. (2020). https://doi.org/10.1016/j.ijid.2020.05.045
ECDC (2020) Guidance for discharge and ending isolation in the context of widespread community transmission of COVID-19. https://www.ecdc.europa.eu/sites/default/files/documents/covid-19-guidance-discharge-and-ending-isolation-first%20update.pdf. Accessed 17 May 2020
V.M. Corman, O. Landt, M. Kaiser, R. Molenkamp, A. Meijer, D.K. Chu, T. Bleicker, S. Brunink, J. Schneider, M.L. Schmidt, D.G. Mulders, B.L. Haagmans, B. van der Veer, S. van den Brink, L. Wijsman, G. Goderski, J.L. Romette, J. Ellis, M. Zambon, M. Peiris, H. Goossens, C. Reusken, M.P. Koopmans, C. Drosten, Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro. Surveill. 25(3) (2020). https://doi.org/10.2807/1560-7917.ES.2020.25.3.2000045
H.G. Muller, J.L. Wang, Hazard rate estimation under random censoring with varying kernels and bandwidths. Biometrics 50(1), 61–76 (1994)
M. Puig-Domingo, M. Marazuela, A. Giustina, COVID-19 and endocrine diseases. A statement from the European Society of Endocrinology. Endocrine 68(1), 2–5 (2020). https://doi.org/10.1007/s12020-020-02294-5
F. Rubino, S.A. Amiel, P. Zimmet, G. Alberti, S. Bornstein, R.H. Eckel, G. Mingrone, B. Boehm, M.E. Cooper, Z. Chai, S. Del Prato, L. Ji, D. Hopkins, W.H. Herman, K. Khunti, J.C. Mbanya, E. Renard, New-onset diabetes in Covid-19. N. Engl. J. Med. (2020). https://doi.org/10.1056/NEJMc2018688
R. Pal, S.K. Bhadada, COVID-19 and diabetes mellitus: an unholy interaction of two pandemics. Diabetes Metab. Syndr. 14(4), 513–517 (2020). https://doi.org/10.1016/j.dsx.2020.04.049
N. Jafar, H. Edriss, K. Nugent, The effect of short-term hyperglycemia on the innate immune system. Am. J. Med. Sci. 351(2), 201–211 (2016). https://doi.org/10.1016/j.amjms.2015.11.011
W. Guo, M. Li, Y. Dong, H. Zhou, Z. Zhang, C. Tian, R. Qin, H. Wang, Y. Shen, K. Du, L. Zhao, H. Fan, S. Luo, D. Hu, Diabetes is a risk factor for the progression and prognosis of COVID-19. Diabetes Metab. Res. Rev. e3319 (2020). https://doi.org/10.1002/dmrr.3319
A.K. Singh, R. Gupta, A. Ghosh, A. Misra, Diabetes in COVID-19: prevalence, pathophysiology, prognosis and practical considerations. Diabetes Metab. Syndr. 14(4), 303–310 (2020). https://doi.org/10.1016/j.dsx.2020.04.004
Y. Fu, P. Han, R. Zhu, T. Bai, J. Yi, X. Zhao, M. Tao, R. Quan, C. Chen, Y. Zhang, Q. He, M. Jing, X. Xiong, D. Tian, W. Yan, Risk factors for viral RNA shedding in COVID-19 patients. Eur. Respir. J. (2020). https://doi.org/10.1183/13993003.01190-2020
Acknowledgements
We thank the Swiss National Science foundation, the Bangerter-Rhyner Foundation, and the Ente Ospedaliero Cantonale (regional public hospitals network, EOC). Open access funding provided by Università della Svizzera italiana.
Funding
N.B. is currently receiving a Mobility grant from the Swiss National Science Foundation (Grant No.: P400PM_183865) and a grant from the Bangerter-Rhyner Foundation. These grants support his fellowship in Paris.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation and analysis were performed by N.B., P.T., V.F.O., V.C., T.M., and E.B. The first draft of the paper was written by N.B. and P.T.; and all authors commented on previous versions of the paper. All authors read and approved the final paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The studies were approved by the regional ethic committee. This study has the following ethic number: 2020-01216 CE 36641.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Buetti, N., Trimboli, P., Mazzuchelli, T. et al. Diabetes mellitus is a risk factor for prolonged SARS-CoV-2 viral shedding in lower respiratory tract samples of critically ill patients. Endocrine 70, 454–460 (2020). https://doi.org/10.1007/s12020-020-02465-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12020-020-02465-4