Abstract
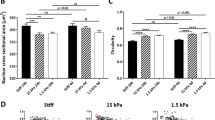
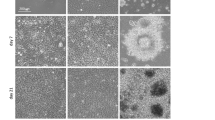
Several studies have demonstrated the possibility to revert differentiation process, reactivating hypermethylated genes and facilitating cell transition to a different lineage. Beside the epigenetic mechanisms driving cell conversion processes, growing evidences highlight the importance of mechanical forces in supporting cell plasticity and boosting differentiation. Here, we describe epigenetic erasing and conversion of dermal fibroblasts into insulin-producing cells (EpiCC), and demonstrate that the use of a low-stiffness substrate positively influences these processes. Our results show a higher expression of pluripotency genes and a significant bigger decrease of DNA methylation levels in 5-azacytidine (5-aza-CR) treated cells plated on soft matrix, compared to those cultured on plastic dishes. Furthermore, the use of low-stiffness also induces a significant increased up-regulation of ten-eleven translocation 2 (Tet2) and histone acetyltransferase 1 (Hat1) genes, and more decreased histone deacetylase enzyme1 (Hdac1) transcription levels. The soft substrate also encourages morphological changes, actin cytoskeleton re-organization, and the activation of the Hippo signaling pathway, leading to yes-associated protein (YAP) phosphorylation and its cytoplasmic translocation. Altogether, this results in increased epigenetic conversion efficiency and in EpiCC acquisition of a mono-hormonal phenotype. Our findings indicate that mechano-transduction related responsed influence cell plasticity induced by 5-aza-CR and improve fibroblast differentiation toward the pancreatic lineage.






Similar content being viewed by others
References
Bird, A. (2002). DNA methylation patterns and epigenetic memory. Genes & Development, 16(1), 6–21.
Palii, S. S., Van Emburgh, B. O., Sankpal, U. T., Brown, K. D., & Robertson, K. D. (2008). DNA methylation inhibitor 5-Aza-2′-deoxycytidine induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyltransferases 1 and 3B. Molecular and Cellular Biology, 28(2), 752–771.
Taylor, S. M., Constantinides, P. A., & Jones, P. A. (1984). 5-Azacytidine, DNA methylation, and differentiation. Current Topics in Microbiology and Immunology, 108, 115–127.
Pennarossa, G., Maffei, S., Campagnol, M., Tarantini, L., Gandolfi, F., & Brevini, T. A. (2013). Brief demethylation step allows the conversion of adult human skin fibroblasts into insulin-secreting cells. Proceedings of the National Academy of Sciences of the United State of America, 110(22), 8948–8953.
Pennarossa, G., Maffei, S., Campagnol, M., Rahman, M. M., Brevini, T. A., & Gandolfi, F. (2014). Reprogramming of pig dermal fibroblast into insulin secreting cells by a brief exposure to 5-aza-cytidine. Stem Cell Reviews, 10(1), 31–43.
Brevini, T. A., Pennarossa, G., Rahman, M. M., et al. (2014). Morphological and Molecular Changes of Human Granulosa Cells Exposed to 5-Azacytidine and Addressed Toward Muscular Differentiation. Stem Cell Reviews.
Mirakhori, F., Zeynali, B., Kiani, S., & Baharvand, H. (2015). Brief azacytidine step allows the conversion of suspension human fibroblasts into neural progenitor-like cells. Cell Journal, 17(1), 153–158.
Tan, S. J., Fang, J. Y., Wu, Y., Yang, Z., Liang, G., & Han, B. (2015). Muscle tissue engineering and regeneration through epigenetic reprogramming and scaffold manipulation. Scientific Reports, 5, 16333.
Brevini, T. A., Pennarossa, G., Acocella, F., Brizzola, S., Zenobi, A., & Gandolfi, F. (2016). Epigenetic conversion of adult dog skin fibroblasts into insulin-secreting cells. The Veterinary Journal.
Chandrakanthan, V., Yeola, A., Kwan, J. C., et al. (2016). PDGF-AB and 5-Azacytidine induce conversion of somatic cells into tissue-regenerative multipotent stem cells. Proceedings of the National Academy of Sciences of the United State of America.
Manzoni, E. F., Pennarossa, G., deEguileor, M., Tettamanti, G., Gandolfi, F., & Brevini, T. A. (2016). 5-azacytidine affects TET2 and histone transcription and reshapes morphology of human skin fibroblasts. Scientific Reports, 6, 37017.
Wakao, S., Kitada, M., Kuroda, Y., et al. (2011). Multilineage-differentiating stress-enduring (Muse) cells are a primary source of induced pluripotent stem cells in human fibroblasts. Proceedings of the National Academy of Sciences of the United State of America, 108(24), 9875–9880.
Ratajczak, M. Z., Ratajczak, J., Suszynska, M., Miller, D. M., Kucia, M., & Shin, D. M. (2017). A Novel View of the Adult Stem Cell Compartment From the Perspective of a Quiescent Population of Very Small Embryonic-Like Stem Cells. Circulation Research, 120(1), 166–178.
Bhartiya, D. (2017). Do Adult Somatic Cells Undergo Reprogramming or Endogenous Pluripotent Stem Cells get Activated to Account for Plasticity, Regeneration and Cancer Initiation? Stem Cell Reviews, 13(5), 699–701.
Christman, J. K. (2002). 5-Azacytidine and 5-aza-2[prime]-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene, 21, 5483–5495.
Stresemann, C., & Lyko, F. (2008). Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. International Journal of Cancer, 123(1), 8–13.
Engler, A. J., Sen, S., Sweeney, H. L., & Discher, D. E. (2006). Matrix elasticity directs stem cell lineage specification. Cell, 126(4), 677–689.
Evans, N. D., Minelli, C., Gentleman, E., et al. (2009). Substrate stiffness affects early differentiation events in embryonic stem cells. European Cell & Materials, 18, 1–13; discussion 13–14.
Huebsch, N., Arany, P. R., Mao, A. S., et al. (2010). Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate. Nature Materials, 9(6), 518–526.
Gilbert, P. M., Havenstrite, K. L., Magnusson, K. E., et al. (2010). Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science, 329(5995), 1078–1081.
Nam, J., Johnson, J., Lannutti, J. J., & Agarwal, S. (2011). Modulation of embryonic mesenchymal progenitor cell differentiation via control over pure mechanical modulus in electrospun nanofibers. Acta Biomaterialia, 7(4), 1516–1524.
Viale-Bouroncle, S., Vollner, F., Mohl, C., et al. (2011). Soft matrix supports osteogenic differentiation of human dental follicle cells. Biochemical and Biophysical Research Communications, 410(3), 587–592.
Schellenberg, A., Joussen, S., Moser, K., et al. (2014). Matrix elasticity, replicative senescence and DNA methylation patterns of mesenchymal stem cells. Biomaterials, 35(24), 6351–6358.
Kshitiz, Park, J., Kim, P., et al. (2012). Control of stem cell fate and function by engineering physical microenvironments. Integrative Biology (Camb), 4(9), 1008–1018.
Merkwitz, C., Blaschuk, O. W., Schulz, A., et al. (2013). The ductal origin of structural and functional heterogeneity between pancreatic islets. Progress in Histochemistry and Cytochemistry, 48(3), 103–140.
Davis, N. E., Beenken-Rothkopf, L. N., Mirsoian, A., et al. (2012). Enhanced function of pancreatic islets co-encapsulated with ECM proteins and mesenchymal stromal cells in a silk hydrogel. Biomaterials, 33(28), 6691–6697.
Pelham, R. J. Jr., & Wang, Y. (1997). Cell locomotion and focal adhesions are regulated by substrate flexibility. Proceedings of the National Academy of Sciences of the United State of America, 94(25), 13661–13665.
Brevini, T. A., Pennarossa, G., Maffei, S., et al. (2012). Centrosome amplification and chromosomal instability in human and animal parthenogenetic cell lines. Stem Cell Reviews, 8(4), 1076–1087.
Shi, Y., Hou, L., Tang, F., et al. (2005). Inducing embryonic stem cells to differentiate into pancreatic beta cells by a novel three-step approach with activin A and all-trans retinoic acid. Stem Cells, 23(5), 656–662.
Nardone, G., Oliver-De La Cruz, J., Vrbsky, J., et al. (2017). YAP regulates cell mechanics by controlling focal adhesion assembly. Nature Communications, 8, 15321.
Tahiliani, M., Koh, K. P., Shen, Y., et al. (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science, 324(5929), 930–935.
Ito, S., D./‘Alessio, Taranova, A. C., Hong, O. V., Sowers, K., L.C., and Zhang, Y. (2010). Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature, 466(7310), 1129–1133.
Zhu, G., Li, Y., Zhu, F., et al. (2014). Coordination of engineered factors with TET1/2 promotes early-stage epigenetic modification during somatic cell reprogramming. Stem Cell Reports, 2(3), 253–261.
Hemberger, M., Dean, W., & Reik, W. (2009). Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington’s canal. Nature Reviews Molecular Cell Biology, 10(8), 526–537.
Higuchi, S., Watanabe, T. M., Kawauchi, K., Ichimura, T., & Fujita, H. (2014). Culturing of mouse and human cells on soft substrates promote the expression of stem cell markers. Journal of Bioscience and Bioengineering, 117(6), 749–755.
Chowdhury, F., Li, Y., Poh, Y. C., Yokohama-Tamaki, T., Wang, N., & Tanaka, T. S. (2010). Soft substrates promote homogeneous self-renewal of embryonic stem cells via downregulating cell-matrix tractions. PLoS ONE, 5(12), e15655.
Walcott, S., & Sun, S. X. (2010). A mechanical model of actin stress fiber formation and substrate elasticity sensing in adherent cells. Proceedings of the National Academy of Sciences of the United State of America, 107(17), 7757–7762.
Dupont, S., Morsut, L., Aragona, M., et al. (2011). Role of YAP/TAZ in mechanotransduction. Nature, 474(7350), 179–183.
Wada, K., Itoga, K., Okano, T., Yonemura, S., & Sasaki, H. (2011). Hippo pathway regulation by cell morphology and stress fibers. Development, 138(18), 3907–3914.
Zhao, B., Li, L., Wang, L., Wang, C. Y., Yu, J., & Guan, K. L. (2012). Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes & Development, 26(1), 54–68.
Halder, G., Dupont, S., & Piccolo, S. (2012). Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nature Reviews Molecular Cell Biology, 13(9), 591–600.
Lian, I., Kim, J., Okazawa, H., et al. (2010). The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes & Development, 24(11), 1106–1118.
Young, R. A. (2011). Control of the embryonic stem cell state. Cell, 144(6), 940–954.
Beyer, T. A., Weiss, A., Khomchuk, Y., et al. (2013). Switch enhancers interpret TGF-beta and Hippo signaling to control cell fate in human embryonic stem cells. Cell Reports, 5(6), 1611–1624.
Varelas, X. (2014). The Hippo pathway effectors TAZ and YAP in development, homeostasis and disease. Development, 141(8), 1614–1626.
Candiello, J., Singh, S. S., Task, K., Kumta, P. N., & Banerjee, I. (2013). Early differentiation patterning of mouse embryonic stem cells in response to variations in alginate substrate stiffness. Journal of Biological Engineering, 7(1), 9.
Richardson, T., Kumta, P. N., & Banerjee, I. (2014). Alginate encapsulation of human embryonic stem cells to enhance directed differentiation to pancreatic islet-like cells. Tissue Engineering Part A, 20(23–24), 3198–3211.
Jaramillo, M., Singh, S. S., Velankar, S., Kumta, P. N., & Banerjee, I. (2015). Inducing endoderm differentiation by modulating mechanical properties of soft substrates. Journal of Tissue Engineering and Regenerative Medicine, 9(1), 1–12.
Polak, M., Bouchareb-Banaei, L., Scharfmann, R., & Czernichow, P. (2000). Early pattern of differentiation in the human pancreas. Diabetes, 49(2), 225–232.
Piper, K., Brickwood, S., Turnpenny, L. W., et al. (2004). Beta cell differentiation during early human pancreas development. The Journal of Endocrinology, 181(1), 11–23.
Brevini, T. A., Pennarossa, G., Maffei, S., Zenobi, A., & Gandolfi, F. (2016). Epigenetic Conversion as a Safe and Simple Method to Obtain Insulin-secreting Cells from Adult Skin Fibroblasts. Journal of Visualized Experiments, (109).
Acknowledgements
This work was funded by Carraresi Foundation, by European Foundation for the Study of Diabetes (EFSD) and by Ricerca Corrente at Centro Cardiologico Monzino, IRCCS. The Laboratory of Biomedical Embryology is member of the COST Action CA16119 In vitro 3-D total cell guidance and fitness (CellFit), the COST Action BM1308 Sharing advances on large animal models (SALAAM) and the COST Action CM1406 Epigenetic Chemical Biology (EPICHEM). The Authors are indebted with Drs Loredana Casalis and Denis Scaini at ELETTRA-Sincrotrone for helping with assessment of the substrates compliance with AFM-assisted nano-indentation methods.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Pennarossa, G., Santoro, R., Manzoni, E.F.M. et al. Epigenetic Erasing and Pancreatic Differentiation of Dermal Fibroblasts into Insulin-Producing Cells are Boosted by the Use of Low-Stiffness Substrate. Stem Cell Rev and Rep 14, 398–411 (2018). https://doi.org/10.1007/s12015-017-9799-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12015-017-9799-0