Abstract
The trophoblast cell lineage is specified early at the blastocyst stage, leading to the emergence of the trophectoderm and the pluripotent cells of the inner cell mass. Using a double mRNA amplification technique and a comparison with transcriptome data on pluripotent stem cells, placenta, germinal and adult tissues, we report here some essential molecular features of the human mural trophectoderm. In addition to genes known for their role in placenta (CGA, PGF, ALPPL2 and ABCG2), human trophectoderm also strongly expressed Laminins, such as LAMA1, and the GAGE Cancer/Testis genes. The very high level of ABCG2 expression in trophectoderm, 7.9-fold higher than in placenta, suggests a major role of this gene in shielding the very early embryo from xenobiotics. Several genes, including CCKBR and DNMT3L, were specifically up-regulated only in trophectoderm, indicating that the trophoblast cell lineage shares with the germinal lineage a transient burst of DNMT3L expression. A trophectoderm core transcriptional regulatory circuitry formed by 13 tightly interconnected transcription factors (CEBPA, GATA2, GATA3, GCM1, KLF5, MAFK, MSX2, MXD1, PPARD, PPARG, PPP1R13L, TFAP2C and TP63), was found to be induced in trophectoderm and maintained in placenta. The induction of this network could be recapitulated in an in vitro trophoblast differentiation model.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human embryo development begins with the fusion of the female and male gametes, followed by the first cleavage divisions that lead to the formation of an 8-cell embryo in about 3 days [1]. At day 4, the embryo develops into a 16-cell morula, starts compaction and becomes a blastocyst. The inner cell mass (ICM) of the blastocyst will then go through gastrulation and organogenesis, whereas the trophectoderm (TE) cells that form the outer envelope of the blastocyst will give rise to the placenta. The specification of the TE and ICM lineages during blastocyst formation is the first developmental decision during human embryo development [2]. It is a key step that paves the way for all subsequent developmental events. Moreover, the earliest stages of trophoblast differentiation are critical for mediating implantation and fostering normal placental growth and function during gestation. Aberrant trophoblast development is associated with serious complications during pregnancy, including recurrent miscarriages, pre-eclampsia and restricted fetal growth.
In mice, TE identity is already set at the blastocyst stage. Indeed, it is possible to isolate embryonic stem (ES) cells from the ICM [3] and trophoblast stem (TS) cells from the trophectoderm [4]. In mouse, the identification of the determinants of trophoblast cell fate mainly stems from the analysis of targeted mutations [5, 6]. At the top of the transcription factor (TF) cascade involved in TE development lies Tead4, which in turn controls Cdx2, Eomes and Tcfap2c. Further downstream, other TFs that play an important role in normal TE development include Gata3, Elf5 and Ets2. Over-expression of these TFs in ES cells revealed that they can be divided in two functionally different groups, one resulting in the conversion of ES cells into self-renewing TS cells (Tead4, Cdx2, Tcfap2c and Eomes), and the other pushing TS cell differentiation into post-mitotic cells (Elf5, Ets2, Gata3) [5]. Growth factors also play important roles in TE development. For instance, Fgf4, which is secreted by the ICM, is crucial for TE development as over-expression of a dominant negative Fgf receptor precludes TE cell division [7]. Conversely, the Hippo signaling pathway must be suppressed to promote the activity of Tead4 via the nuclear localization of its co-factor YAP [8].
To what extent are these findings relevant to understand human TE development? Conserved features between rodents and humans include the mutually exclusive expression of key TFs in ICM (NANOG, OCT4 and SOX2) and TE (CDX2), as well as the relatively lower DNA methylation of TE in comparison to ICM [5, 9]. But there are also some major differences, such as the failure of deriving human TS cells or the fact that, in humans, the initial implantation contact occurs through the polar TE. Therefore, improving our knowledge about the molecular features of this first developmental cell lineage split in humans is mandatory. Whole embryo microarray experiments have contributed to the description of the first 3 days of the human embryo [10, 11]. Studies in human ES cells (hESC) have opened the way to understanding the ICM transcriptional network [12] as hESC are derived from the ICM and retain its cardinal features (i.e., pluripotency, the core transcriptional regulatory network and intense proliferation) [13–15]. By contrast, knowledge on the transcriptional network that regulates early human trophoblast development is still fragmentary with very few published studies in which few samples were used and only part of the human transcriptome was covered [16].
Here, we report the whole genome transcriptome analysis of five mechanically dissected human mural TE samples. By comparing the TE gene expression profile with that of human pluripotent stem cells (PSCs) and placenta samples, we uncovered a tightly connected network of TFs that are expressed during TE development and in mature placenta. Moreover, induction of this transcriptional network could be recapitulated in vitro by inducing hESC and induced pluripotent stem (iPS) cells to differentiate into trophoblast cells. This new insight into early TE development contributes not only to our knowledge on human development but also to improving assisted reproductive technologies.
Results and Discussion
The Mural Trophectoderm Transcriptome
Mural TE was mechanically separated from the inner cell mass (ICM) of five fresh blastocyst stage (day 5) embryos produced by in vitro fertilization (IVF) and the five TE samples were then individually analyzed by whole genome Affymetrix oligonucleotide microarrays. Validation of the microarray data for three genes that were strongly up-regulated in the TE samples (DNMT3L, GAGE2 and GATA3) was performed by real-time quantitative PCR using five independent mural TE samples (Supplementary Figure S1). Unsupervised hierarchical clustering of the gene expression level data of the five TE samples and of a large panel that included hESC, germ cells, placenta and different adult tissue samples (n = 181) (see Materials and Methods and Supplementary Table S1) divided the samples in three main branches: a first one containing all the nervous system samples; a second one that included the TE, testis/oocytes, hESC, fibroblast and placenta samples, and a third branch containing all the other adult tissues (Fig. 1a). As expected, the TE samples clustered together with samples that were developmentally closer to them.
Overview of TE expression profile and expression of cell cycle-specific genes in TE, hESCs, placenta and nervous system. a Unsupervised hierarchical clustering of the 181 panel samples. The first 30000 PS with the highest coefficient variation were analyzed with the Cluster software. Three cluster branches emerged: the nervous system branch (green), embryonic development and gamete branch (pink) and adult tissue branch (blue). b Proportion of genes from the cell cycle signature that are present in the TE, hESC, placenta and nervous system gene signatures
Samples in the TE branch all shared a strong cell cycle gene signature. Indeed, many genes that regulate cell division and cell proliferation, such as CDK1/CDC2, MCM7 and NMYC, were highly expressed in TE, demonstrating a strong cell cycle activity. To assess the proportion of cell cycle genes expressed in TE, we built a cell cycle signature, following a strategy that we previously used to monitor the expression profile of cell cycle genes in hESCs (see Materials and Methods). We then compared the proportion of these “cell cycle” probesets (PS) (see Supplementary Table S2 for the PS/gene list) present in the hESC, TE, placenta and adult nervous system gene expression signatures. The nervous system group was included as a negative control because neuronal cells do not (or very little) proliferate. We found that hESC and TE samples included respectively 28.5% and 16.7% of the cell cycle PS, whereas the placenta contained only 3.1% and the nervous system group none (Fig. 1b). These results demonstrate an intense cell proliferation activity in the five TE samples. Indeed, like hESCs will give rise to the fetus through massive cell expansion, TE cells also need to considerably expand to contribute to the development of the placenta, a temporary organ of about 500 grams.
Comparison with hESCs: The Human Trophectoderm Expression Signature
In order to better delineate the first irreversible developmental cell lineage divergence that takes place during blastocyst formation, we compared the five mural TE samples to ten hESC lines (see Supplemental Table S3 for detailed information). By applying a significance analysis of microarray (SAM) with a false discovery rate (FDR) <0.5% and a fold change of at least 5, followed by filtering based on the expression call, we delineated a ≪TE signature≫ of 975 PS that are over-expressed in TE (Supplementary Table S4) and a ≪hESC signature≫ of 1018 PS that are up-regulated in hESC (Fig. 2a). The hESC signature included known pluripotency genes, such as OCT4/POU5F1, NANOG, LEFTY 1 and 2, THY1 and FDZ7 [14]. The pluripotency gene LIN28, which represses the let-7 miRNA and is preferentially expressed in hESCs and oocytes, was not present in the hESC signature because it is also highly expressed in TE cells. Similarly, ZFP42, the human homolog of mouse Rex, which is strongly associated with pluripotency [17], was simultaneously over-expressed in hESCs and TE, and thus was excluded from the hESC signature. Conversely, the TE signature was characterized by the expression of genes, such as CGA, PGF and ALPPL2, that are involved in the functions of the human placenta [18] as well as of genes that are important for mouse placental development, such as PPARG [19] (Fig. 2b). CDX2 expression was not detected by the microarrays possibly because its expression level was below the sensitivity of the microarray RNA amplification technique. Alternatively, it could be explained by a regional CDX2 gradient with a lower expression in mural than in polar TE. This hypothesis is supported by the heterogeneous expression of CDX2 in mouse extra-embryonic tissues [5]. Overall, the TE and the hESC signatures are in agreement with what is known about TE and hESC biology.
To further characterize the TE signature, we identified the gene ontology (GO) functional categories that are over- or under-represented in the TE signature. In comparison to the hESC signature, the TE signature was significantly enriched in genes implicated in cell protein synthesis, such as genes related to the categories ribosome (GO:0005840), structural constituent of ribosome (GO:0003735), biosynthetic process (GO:0009058), and translation (GO:0006412) (Fig. 3a and Supplementary Figure S2). The TE enrichment for genes involved in protein synthesis was remarkable and may in part be explained by CGA secretion as suggested by the high level of CGA mRNA. Conversely, the hESC signature was characterized by enrichment in genes involved in regulation of development and cell differentiation, such as anatomical structure development (GO:0048856), multicellular organismal development (GO:0007275) and cell adhesion (GO:0007155) (Fig. 3a and Supplementary Figure S2). Accordingly, in the hESC samples, developmental TFs, such as OTX2, HEY2, PBX1, were significantly up-regulated. The expression pattern of these TFs suggests that their expression in hESCs is not the manifestation of partial differentiation of such cells in culture, but rather an intrinsic property of these cells. Indeed, OTX2, a homeodomain-containing TF involved in brain and sensory organ development, and HEY2, a basic-loop-helix factor known to be involved in arterial-venous cell fate decision, are already expressed at the oocyte stage, before fertilization (see Supplementary Figure S5), suggesting a rapid and specific silencing in TE cells, whereas their expression is conserved in hESCs. Similarly, PBX1, a TF that control limb development from Drosophila to human, IRX1 and IRX3, two members of the Iroquois homeobox gene family, which play multiple roles during pattern formation of vertebrate embryos, were found expressed in many tissues but not in TE, suggesting a specific silencing in TE cells.
Gene Ontology analysis of the TE and hESC signatures. a A Gene Ontology (GO) term enrichment analysis was carried out using FatiGO+. Only the GO terms that are significantly different (using adjusted p-values) between the two signatures are shown. A tag cloud shows the significant GO terms in each signature. The character size of each tag is proportional to their significance (see Materials and Methods). The details and the adjusted p-value of each term are shown in Supplementary Figure S2. b Heat map of the expression of all the extracellular matrix genes tagged by the GO term: 0031012 in the TE and hESC signatures showing the strong differential expression of extracellular matrix gene families in TE and hESC samples
Another striking divergence between TE and hESCs was the differential expression of genes coding for proteins that were related to GO cellular component categories, such as “extracellular space” and/or “extracellular matrix”, especially Collagens and Laminins as shown by the heat map of the TE and hESC signatures in which genes that share the GO annotation “extracellular matrix” (GO0031012) are highlighted (Fig. 3b). The TE signature contained 4.7 folds less “extracellular matrix” PS than the hESC signature. To exclude the possibility that the Collagen gene up-regulation observed in the hESC signature was caused by fibroblast feeder contamination, we added transcriptome data about two feeder-free hESC samples (see Supplemental Table S3) into the heat map, and confirmed that hESCs express many Collagen genes in contrast to the TE samples (Fig. 3b). Conversely the TE signature was significantly enriched in Laminin genes (LAMA1, A5, B1 and C1). These observations are in agreement with the expression of Laminin 111 and 511 in rodent blastocysts [20]. These new results also bring an answer to the issue of the origin of the basal lamina lining the human blastocoel cavity, demonstrating that mural TE is a major source of Laminins, a key component of the basal lamina [21, 22]. Human ESC and iPS cells depend on extracellular matrix components for their growth, including Laminin (the major component of the basement membrane matrix Matrigel). Since Laminin genes are expressed in TE cells but not in the ICM, we cannot exclude that ICM cells depend for their growth on the Laminins produced by the TE cells. We previously showed that hESCs express significantly less genes coding for extracellular compartment proteins than differentiated tissues [14] and that oocytes express these genes at a lower level than cumulus cells, in agreement with the fact that oocytes are solitary cells that are not embedded in a tissue [23]. Altogether, these results suggest that extracellular matrix genes are globally silent before fertilization, then they are partially expressed in hESCs, but mostly not in TE (except for Laminins), and finally they become strongly expressed as soon as somatic cell differentiation begins.
Another remarkable gene of the TE signature is ABCG2, a membrane transporter of the ATP-binding cassette (ABC) super-family. This gene is involved in the efflux of xenobiotics with a well-documented role in the resistance of malignant cells to anti-cancer drugs [24]. In addition, high expression of ABCG2 in specific stem cell subpopulations makes it possible to purify “side cell populations”, including hESCs, based on the efflux of the Hoechst 33342 dye [25, 26]. ABCG2 is also expressed and functional in placenta, possibly to protect the fetus [27, 28]. Strikingly, ABCG2 was expressed at a very high level in TE cells, 90-fold higher than in hESCs and 7.9-fold higher than in placenta (Supplemental Figure S5), which may reflect the distinct susceptibility of the early embryo to xenobiotics and suggests a major role of this gene in its protection.
Cancer/Testis Antigen Expression in Trophoblast and the Epigenetic Pattern in TE
Another feature of the TE signature was the up-regulation of several cancer/testis (CT) antigens. C/T gene expression was first described in various cancers and in testis. More recent studies have reported expression in other normal tissues, such as pancreas, liver, spleen and placenta [29, 30]. Their function remains poorly documented, although a role in transcription activation or repression and, for some of them, more specifically in gametogenesis has been described [29]. Recent reports have shown that some C/T genes are expressed during human early embryonic development and in placenta [31–33]. Here we report a strong expression in TE of several C/T antigen genes from the GAGE, MAGE, PAGE and XAGE families, all located on the X chromosome (CT-X genes) (see Supplementary Figure S5). The most over-expressed C/T gene was GAGE3, with an expression 1155-fold higher in TE than in hESC samples. Other GAGE family members were also highly detected, but due to the high sequence conservation among the members of this family, they could not be precisely identified by microarray analysis. Although the expression of these C/T genes in TE may be linked to a functional role, their up-regulation could also be an indirect consequence of the genome hypomethylation of TE cells [9]. Indeed, CT-X gene expression is reported to be correlated with the methylation level of their promoters and their expression can be induced by experimental chromatin demethylation even in cells that do not normally express them [29, 34, 35].
Trophectoderm-Specific Genes
To establish a list of TE-specific genes, we compared the transcriptome data of the five TE samples to the data of all the other samples of the panel (n = 176). A SAM analysis with a FDR <0.01% and fold change >5 revealed that 978 PS were significantly up-regulated in TE compared to the other tissues. Within these PS, 18 PS displayed a signal 10 times stronger in TE than in any other tissue and were thus considered “TE-specific” (Table 1). Some selected genes are illustrated in Fig. 4. This tissue selectivity strongly suggests a role for these genes in the development and function of human mural TE. The specific expression of Alkaline Phosphatase Placental-Like 2 (ALPPL2) is in agreement with the reported expression of this gene in the placenta lineage. However, two different PS displayed very contrasting expression patterns (Supplemental Figure S3). One PS located in the 5′ region of ALPPL2 (exons 3 and 4) was highly expressed in placenta samples and weakly in TE samples. The other PS, located in the 3′ untranslated region of ALPPL2 was specifically expressed in TE samples, suggesting a splicing event determined by the developmental stage of the placental tissue. Laminin alpha 1 (LAMA1) was another TE-specific gene, thus further supporting the hypothesis of a major role for Laminins during early human embryo development, especially in compartmentalization via the basement membrane and in orienting the direction and quality of trophoblast invasion similarly to what observed in rodents [20, 36]. The very strong, TE-specific expression of CCKBR was unexpected. There are two types of human Cholecystokinin (CCK) receptors. The B receptor (CCKBR) shows high affinity for Gastrin and CCK. Studies in the mouse showed that it is involved in digestion stimulation and regulation of neurotransmitter levels, but also in cell proliferation [37], migration [38] and invasion [39]. Wroblewski et al. have reported that the CCKBR pathways induce the expression of Matrix Metalloproteinase 9 (MMP9) [39], but we could not find any over-expressed MMP in the TE signature. Since our TE samples were derived from pre-implantation embryos, CCKBR might be involved in readying the trophoblast for invasion, whereas the invasion machinery will be operational only after contact with the endometrium. Overall, many TE-specific genes have no known function (see Table 1), reflecting the fact that human TE is poorly studied and that the most TE-specific genes have been overlooked or may not have important roles in other, more accessible tissues. The finding that many receptors, including CCKRB, are expressed in human TE cells, provides new opportunities for understanding the relationships between TE and ICM during placenta differentiation and for using their expression in TE as biomarkers for blastocyst selection. We can also speculate that this new knowledge might provide new strategies for therapeutic interventions on the developing placenta either by blocking or stimulating these receptors.
Expression profile of five TE-specific genes in the panel of samples. The specific over-expression of ALPPL2, KHDCL1, CCKBR, DNMTL3 and LAMA1 in TE samples in comparison to all the other embryonic or adult samples of the panel is illustrated by bar graphs obtained using the Amazonia! gene atlas explorer (http://www.amazonia.transcriptome.eu). OCT4, a pluripotency gene, and ACTG1, a housekeeping gene, are also included
Selective Expression of DNMT3L in Trophectoderm
The expression of DNA (cytosine-5-)-Methyltransferase 3-Like (DNMT3L) in TE was 371 fold higher than in all the other tissues, including adult germinal tissues. DNMT3L encodes a nuclear protein that is similar to DNA methyltransferases but lacks the domain coding for the methyltransferase activity and thus is a non-functional homologue of DNA methyltransferases. In mouse, DNMT3L is an important regulator of the activity of the DNA methyltransferases DNMT3A and DNMT3B and plays an essential role in the establishment of maternal genomic imprints [40] and de novo DNA methylation [41, 42]. The absence of DNMT3L expression in oocytes before fertilization and in pluripotent stem cells and its considerable expression in TE cells could be explained by a lineage-specific expression of this DNA methyltransferase. However, DNMT3L mRNA was completely absent also in placenta samples (Fig. 4). A precise and limited window of expression is a recurrent feature of DNMT3L. Indeed, in mouse gametes, DNMT3L is only expressed in growing oocytes and in non-dividing prospermatogonia until the time of birth with a complete extinction by day 6 post-partum [43]. Our findings add a third tissue with a transient burst of DNMT3L expression, strongly suggesting a functional role in the placenta lineage during its very early developmental steps. DNMT3L plays a crucial role in gene imprinting and retrotransposon control and may also play a role in regulating DNA methylation in TE.
A Core Transcriptional Network from Trophectoderm to Placenta
To better understand the molecular determinants of the TE and ICM lineage segregation, we focused on the TFs that are over-expressed in TE but not in hESC. The TE signature comprised 64 PS coding for 51 TFs, including genes like GCM1, NR6A1/RTR and PPARG that are important for placental development [19, 44, 45]. We hypothesized that if some of these TFs were necessary to instruct and maintain the TE lineage throughout placenta development, they would not only be preferentially expressed in TE and not in hESCs, but their expression would also be maintained until completion of placenta development. We thus computed a placenta signature by comparing the six placenta samples to the ten hESC samples. The placenta signature contained 2328 PS in which 143 PS represented genes coding for TFs. By intersecting the TFs of the TE and the placenta signatures, we found 16 TFs (Fig. 5a and Table 2) that were expressed both in TE samples (when the TE lineage emerges in blastocysts) and in mature placenta. This specific expression profile suggests that these 16 TFs play a critical role in establishing and maintaining the trophoblast lineage. Some of these 16 TFs have already been reported to be important for trophoblast differentiation or placenta formation. For instance, GCM1, a placenta-specific TF that is necessary for placental development [46], was robustly expressed in the five TE samples and the six placenta samples (Supplementary Figure S5). GCM1 induces ERVWE1 expression in trophoblast, where endo-retroviruses (ERV) cause cell fusion and contribute to form the syncytium structure [47, 48]. GATA-2 and GATA-3 are known to be expressed in mouse TS lines and in placenta and to regulate placenta-specific genes [49]. Finally, Peroxisome Proliferator-Activated Receptor Gamma (PPARG), a nuclear hormone receptor implicated in the differentiation of various cell types, is necessary for trophoblast and placental differentiation in mice because PPARG-null mice die at E10 due to defects in these tissues [19].
A core transcription factor (TF) network is over-expressed in TE samples and maintained also in mature placenta samples. a Comparison of the placenta and TE transcriptomes to the hESC transcriptome identified 16 transcription factors (TFs) that are over-expressed in both TE and placenta. b Among these 16 TFs, a network of 13 TFs (in orange) was found with the help of IPA (Ingenuity). The size of the circles representing the TFs is proportional to their fold change in expression between TE and hESCs. The genes in grey were added by IPA to form the network: these genes are expressed in TE and placenta, but without being significantly over-expressed. Four genes in this network are induced by BMP4 according to the IPA analysis
We then analyzed the functional relationship between these 16 TFs using the Ingenuity Pathway Analysis (IPA) software. Remarkably, 13 of the 16 TFs displayed a documented functional interaction with each other, forming a tightly connected network (Fig. 5b). A detailed list of the documented interactions of theses TFs is in Supplementary Figure S4 and Supplementary Table S5. Given that several of these 13 TFs were known to be important in the trophoblast cell lineage specification and because of the important connectivity, this network was therefore designed as the “TE core transcriptional regulatory circuitry”. Most of these interactions were described in cell types other than TE (for instance, CEBPA induction by PPARG in activated hepatic stellate cells) [50], but the concomitant and strong expression of these TFs in the TE samples supports the hypothesis that this functional cross-talk also occurs in TE.
Induction of the Trophectoderm Transcription Factor Network During In Vitro Trophectoderm Differentiation of Pluripotent Stem Cells
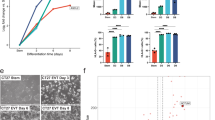
To substantiate the TE core transcriptional regulatory circuitry induction in the trophoblast lineage, we chose to challenge this network in an in vitro model of trophoblast development. Human hESCs differentiate into extra-embryonic lineages, including trophoblast cells, when cultured in the presence of BMP4 [51, 52]. Consistent with previously published results, the hESC lines H1 and HD83 and the iPS cell line M4C2 underwent morphological changes (flattened cell shape, cobblestone appearance and patches of cells expressing GATA3) from day 3 after BMP4 addition (Fig. 6a and b). In some colonies, syncytium-like structures appeared after one to two weeks (Fig. 6a). Trophoblast markers, such as CGA, CDX2 and KRT18, were induced in the BMP4-differentiating cell population as early as day 5 after BMP4 addition (Fig. 6c). In this model, we tested the expression of nine TFs of the TE core transcriptional regulatory circuitry. Indeed, we tested nine players of this network and found that eight of them were significantly induced (GATA2, GATA3, GCM1, TB63, TFAP2C, CEBPA, PPP1R13L and PITX2) (p < 0.05) during in vitro TE differentiation (Fig. 6d). Four genes of this network are known to be induced by BMP4 (Fig. 5b) and since BMP4 is essential for in vitro trophoblast differentiation of hESC [51], it could therefore play an upstream role in inducing the complete network. Collectively, these data strongly suggests that these TFs form a TE core transcriptional regulatory circuitry that plays an active role in the induction and maintenance of the gene expression program of the placental lineage. This provides a new tool to better understand the causes of early miscarriage linked to defects in the development of trophectoderm and placenta. It would be also important to investigate whether the genes of the TE core transcriptional regulatory circuitry are differentially expressed in the developing placenta in cases of spontaneous miscarriage relative to cases of therapeutic pregnancy termination.
Induction of the TE core transcriptional circuitry induction is recapitulated during in vitro differentiation of pluripotent stem cells into trophoblast cells. a Trophoblast differentiation of pluripotent stem cells was induced by adding BMP4. Morphological changes of the hESC line HD83 cultured on Matrigel in MEF-conditioned medium after 3 days in the presence of 10 ng/mL FGF2 (left panel, negative control) or 10 ng/mL BMP4 (middle panel), and after 12 days with 10 ng/mL BMP4. Scale bar is 50 μm. b Immunofluorescence analysis showing the nuclear expression of GATA3 in HD83 cells after 5 days with BMP4. Scale bar is 10 μm. c After 5 days in the presence of 10 ng/mL BMP4, the hESC line HD83 and the iPS cell line M4C2 displayed increased expression of known trophoblast markers (CGA, KRT18 and CDX2) and decreased expression of the pluripotency marker OCT4. Expression changes were calculated by normalizing the gene expression first to the expression of the housekeeping gene GAPDH and then to gene expression in FGF2-treated control cells. * p < 0.05 and dashed line indicates the 1 fold change level. d The expression of GATA2, GATA3, GCM1, TB63, TFAP2C, CEBPA, PPP1R13L and PITX2 (TE core transcriptional regulatory circuitry) was significantly up-regulated in differentiated hESC and iPS cells (5 days with BMP4). * p < 0.05 and dashed line indicates the 1-fold change level
Conclusion
The trophoblast cell lineage is essential for the development of mammalian embryos in utero. It is however, poorly studied at its earliest stage in humans, due to the scarcity of material. Therefore, our comparison of five mural trophectoderm transcriptomes with those of pluripotent cells, oocytes, placenta and different adult tissues provides new data on the transcriptional properties of this tissue in human. The description of several remarkable features of trophectoderm, such as the specific expression of Laminins, Cancer/Testis genes, CCKBR and DNMT3L, opens new directions for understanding trophoblast cell lineage development but also implantation disorders. Most remarkably, the identification of a TE core transcriptional regulatory circuitry, which is maintained until the placenta stage, provides a new framework for delineating the molecular control of the TE lineage. These results open new avenues for medical interventions by providing a better understanding of the molecular causes of early miscarriages and for improving the implantation rate by modulating the level of the ligands of the receptors expressed in TE cells.
Materials and Methods
Trophectoderm Preparation
Mural TE samples were mechanically separated from the ICM of individual blastocysts under an inverted microscope (Leica, Germany), then lysed in RLT RNA extraction buffer and frozen at −80°C. Blastocysts were donated for research after informed consent by couples for our hESC cell derivation project approved by the Agence de la Biomédecine. Overall, we successfully derived five hESC lines, but none of them originated from the embryos from which the TE samples were isolated. Five TE samples from blastocysts of two patients were used for the microarray analyses and five additional TE samples from blastocysts of three other patients were used for qRT–PCR validation of the microarray data.
Microarray Data Collection and Normalization
Total RNA was purified from the TE samples using the RNeasy Micro kit (Qiagen, Courtaboeuf, France) with an additional RNAse-Free DNase step to eliminate residual DNA, and quantified with a NanoDrop spectrophotometer (Thermo Fischer,Wilmington, Delaware, USA). A double amplification of total RNA was used to generate suitable quantity of labeled cRNA for hybridization to U133 plus 2.0 GeneChip pangenomic oligonucleotide arrays (Affymetrix, Santa Clara, CA, USA) as previously described [53]. We recently reported a comparison of these five TE microarray data with human endometrium [54].
In parallel, we collected U133 Plus 2.0 chip microarray data from published reports or from our laboratory and assembled a panel of 181 samples that includes transcriptome data on hESC, oocyte, placenta, different adult tissues and the five TE samples. In addition, transcriptome data for another 28 non-proliferating and 36 highly proliferating tissue samples, which are independent from the panel, were used for the cell cycle analysis and data about two other hESC samples from feeder-free cultures were collected for the extracellular matrix analysis. These data are available in the Gene Expression Omnibus (GEO) database and the list of these samples with their GEO accession number is in Supplementary Tables S1 and S3. All microarray data were normalized and analyzed using Expression Console (Affymetrix) and the MAS5 algorithm with the default analysis setting and global scaling as first normalization method, with a trimmed mean target intensity value (TGT) of each array arbitrarily set at 100. For each signal measurement, the MAS5 algorithm provides a “detection call”. The ‘call’ is tagged as ‘present’ when the perfect match probes of the probeset (PS) are significantly (p-value <0.05) more hybridized than the mismatch probes; ‘absent’ when both perfect match and mismatch probes display a similar fluorescent signal; and ‘marginal’ when the probeset does not match with the ‘present’ or to the ‘absent’ call criteria.
Data Analysis and Visualization
Hierarchical clustering was carried out with CLUSTER and visualized with the help of TREEVIEW [55]. The whole-genome, unsupervised clustering was run with the 10,000 PS that had the most significant coefficient of variation (CV) of signal across the samples.
Lists of the genes differentially expressed between two classes of samples were obtained using two-class Significance Analysis of Microarray (SAM) (http://www-stat.stanford.edu/~tibs/SAM/) with Wilcoxon test statistic and sample-label permutation (n = 300) [56]. Using this method, we compared the transcriptome data of the five TE samples with data for the ten hESC samples collected from public and in-house databases. With a false discovery rate (FDR) of 0.277%, 7521 PS were found to have a TE/hESC signal ratio ≥ 5 and 975 of these PS (“TE signature”) displayed a “present” call in at least three of the five TE samples. Conversely, 1083 PS were significantly over-expressed in the ten hESC samples compared with the five TE samples with a hESC/TE signal ratio ≥ 5; 1018 of these PS (“hESC signature”) displayed a “present” call in at least three hESC samples. The same method and thresholds were used to compare placenta and hESC samples with an FDR of 0.188%. The parameters and results of each SAM analysis are summarized in Supplementary Table S6. All the TFs in the placenta and TE signatures were defined using Gene Ontology annotations (http://www.geneontology.org/).
For the cell cycle gene analysis, public Human Genome U133 Plus 2.0 chip (Affymetrix) data about 36 highly proliferating (“cycling”) and 28 differentiated adult tissue samples with little proliferation activity (“non-cycling”) (see Supplementary Table S1) were compared using SAM with a FDR <0.2% and a ratio of 5. A group of 664 PS (representing 531 genes), termed “cell cycle signature”, was significantly over-expressed in the “cycling” samples compared to the “non-cycling” samples. The cell cycle signature was intersected with the hESC, TE, placenta and nervous system signatures that were computed using SAM by comparing each of these groups (hESC: n = 10, TE: n = 5, placenta: n = 6 and nervous system: n = 45 samples) with all the other samples of the panel (n = 115).
To compute a list of TE-specific genes, we carried out a two-class, unpaired SAM analysis of the TE samples and the panel samples (n = 176), which identified 978 PS differentially expressed in TE samples (see Supplementary Table S6 for parameters). Of these PS, only those with a “present” call in 3 out of 5 TE samples, a signal value >50 and a mean signal 10-fold higher in TE than in any of the other 23 tissue categories (see the microarray sample list, Supplementary Table S1) were designed as “TE-specific”.
The Gene Ontology annotation analysis was carried out using the FatiGO + tool (BABELOMICS v3.1; http://babelomics.bioinfo.cipf.es) [57]. Only annotations with a false discovery rate-adjusted P-value below 0.05 were considered significant.
The gene expression profile of TE and of the other human tissues can be accessed using our free web interface Amazonia! that allows an easy query of public human transcriptome data by key words in thematic pages (http://amazonia.transcriptome.eu) (Le Carrour et al 2010). The Amazonia! tool was used to generate the bar graphs shown in the figures.
Cell Culture
The human ESC line HD83 was derived in our laboratory and H1 was imported from WiCell [53, 58]. The human induced pluripotent stem cell line M4C2 was reprogrammed by using lentiviral vectors containing the human OCT4/POU5F1, SOX2, NANOG and LIN28 genes [59] and displays all the features of pluripotent stem cells (PSC): M4C2 grow as typical PSCs, are positive for OCT4 and ABCG2 expression and for phosphatase alkaline activity, display a typical PSC expression signature by microarray analysis and can differentiate into cells of all three germ layers (Dijon-Grinand et al, submitted). PSC lines were maintained on irradiated (40 Gy) human foreskin fibroblast feeders [60], in 80% KO-DMEM, 20% KOSR, 2 mM L-glutamine, 1% non-essential amino acids, 0.5 mM β-mercaptoethanol (all from Gibco Invitrogen, Cergy-Pontoise, France) and complemented with 10 ng/mL bFGF (Abcys, Paris, France). PSC were mechanically passaged weekly.
Before trophoblast differentiation, PSC were placed in feeder-free culture conditions: cells were dissociated with 1 mg/mL collagenase IV at 37°C for 20 min, then plated on a 1/30 diluted Matrigel-coated support and cultured in mouse embryonic fibroblast (MEF) conditioned medium with 10 ng/mL of FGF2. For in vitro trophoblast differentiation, MEF-conditioned medium was complemented with 10 ng/mL of BMP4 (R&D System) instead of FGF2. Cells were either harvested for real-time quantitative PCR or fixed for immunofluorescence at day 5, or maintained in culture until week 2 for morphological observations.
To produce MEF-conditioned medium, MEFs were plated in 6-well plates at a concentration of 100 000 cells/mL with 2 mL/well of pluripotent stem cell culture medium without FGF2 and supernatant was collected every day for 1 week.
Reverse Transcription (RT) and Real-Time Quantitative PCR (qPCR)
One μg of total RNA was used for RT in a final volume of 20 μL with the SuperScript® First-Strand Synthesis System (Invitrogen) according to the manufacturer’s instructions. qPCR was performed in 384-well plates (Sorenson BioScience, Inc.) on a Lightcycler ® 480 Real-Time PCR System (Roche Diagnostics). cDNA (1/20 dilution) was added to a reaction mix (final volume of 10 μL) containing 2 μL diluted cDNA, 5 μL Sybr Green (Roche Diagnostics), 0.5 μM forward and reverse primers (primer sequences are listed in supplementary Table S7) and amplification carried out according to the following conditions: 10 min at 95°C, then 55 cycles of 10 s at 95°C, 20 s at 62°C and 25 s at 72°C. At the end, a melting curve from 95°C to 62°C was performed to control primer specificity. GAPDH was used as endogenous control for calculating the relative expression level of each gene, and the gene expression of BMP4-treated cells was normalized to control FGF2-treated cells to compute the expression change.
For the qPCR validation of the microarray data, five independent TE and hESC mRNA samples were amplified using the microarray double in vitro transcription method, followed by qPCR.
Immunofluorescence
After 5 days of BMP4 treatment, PSCs were washed with PBS, fixed in 4% paraformaldehyde and permeabilized with 0.1% Triton-X for 30 min. They were then blocked with 5% goat serum at room temperature for 30 min, incubated with anti-GATA3 (R&D system) antibody (1/20 dilution) and immunolabeling was revealed by incubation with rabbit anti-mouse IgG antibody coupled with Alexa fluor 568 (Molecular Probes) (1/300 dilution) for 1 h. Cell nuclei were detected with DAPI staining. Images were taken with the Isis Fluorescence Imaging System (MetaSystems Germany).
References
Wong, C. C., Loewke, K. E., Bossert, N. L., et al. Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nature Biotechnology, 28, 1115–1121.
Arnold, S. J., & Robertson, E. J. (2009). Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nature Reviews. Molecular Cell Biology, 10, 91–103.
Evans, M. J., & Kaufman, M. H. (1981). Establishment in culture of pluripotential cells from mouse embryos. Nature, 292, 154–156.
Tanaka, S., Kunath, T., Hadjantonakis, A. K., Nagy, A., & Rossant, J. (1998). Promotion of trophoblast stem cell proliferation by FGF4. Science, 282, 2072–2075.
Senner, C. E., & Hemberger, M. (2010). Regulation of early trophoblast differentiation—lessons from the mouse. Placenta, 31, 944–950.
Rossant, J., & Cross, J. C. (2001). Placental development: lessons from mouse mutants. Nature Reviews Genetics, 2, 538–548.
Chai, N., Patel, Y., Jacobson, K., McMahon, J., McMahon, A., & Rappolee, D. A. (1998). FGF is an essential regulator of the fifth cell division in preimplantation mouse embryos. Developmental Biology, 198, 105–115.
Nishioka, N., Inoue, K., Adachi, K., et al. (2009). The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Developmental Cell, 16, 398–410.
Santos, F., Hendrich, B., Reik, W., & Dean, W. (2002). Dynamic reprogramming of DNA methylation in the early mouse embryo. Developmental Biology, 241, 172–182.
Dobson, A. T., Raja, R., Abeyta, M. J., et al. (2004). The unique transcriptome through day 3 of human preimplantation development. Human Molecular Genetics, 13, 1461–1470.
Assou, S., Boumela, I., Haouzi, D., et al. (2011). Dynamic changes in gene expression during human early embryo development: from fundamental aspects to clinical applications. Human Reproduction Update, 17, 272–290.
Boyer, L. A., Lee, T. I., Cole, M. F., et al. (2005). Core transcriptional regulatory circuitry in human embryonic stem cells. Cell, 122, 947–956.
Smith, A. G. (2001). Embryo-derived stem cells: of mice and men. Annual Review of Cell and Developmental Biology, 17, 435–462.
Assou, S., Le Carrour, T., Tondeur, S., et al. (2007). A meta-analysis of human embryonic stem cells transcriptome integrated into a web-based expression atlas. Stem Cells, 25, 961–973.
Ramirez, J., Bai, Q., Dijon-Grinand, M., et al. (2010). Human pluripotent stem cells: from biology to cell therapy. World Journal of Stem Cells, 2, 24–33.
Adjaye, J., Ralf Herwig, H. J., BenKahla, A., Brink, T. C., Wierling, C., Hultschig, C., et al. (2005). Primary differentiation in the human blastocyst: comparative molecular portraits of inner cell mass and trophectoderm cells. Stem Cells, 23, 1514–1525.
Ben-Shushan, E., Thompson, J. R., Gudas, L. J., & Bergman, Y. (1998). Rex-1, a gene encoding a transcription factor expressed in the early embryo, is regulated via Oct-3/4 and Oct-6 binding to an octamer site and a novel protein, Rox-1, binding to an adjacent site. Molecular and Cellular Biology, 18, 1866–1878.
Sood, R., Zehnder, J. L., Druzin, M. L., & Brown, P. O. (2006). Gene expression patterns in human placenta. Proceedings of the National Academy of Sciences of the United States of America, 103, 5478–5483.
Barak, Y., Nelson, M. C., Ong, E. S., et al. (1999). PPAR gamma is required for placental, cardiac, and adipose tissue development. Molecular Cell, 4, 585–595.
Aszodi, A., Legate, K. R., Nakchbandi, I., & Fassler, R. (2006). What mouse mutants teach us about extracellular matrix function. Annual Review of Cell and Developmental Biology, 22, 591–621.
Timpl, R., Rohde, H., Robey, P. G., Rennard, S. I., Foidart, J. M., & Martin, G. R. (1979). Laminin–a glycoprotein from basement membranes. Journal of Biological Chemistry, 254, 9933–9937.
Thorsteinsdottir, S. (1992). Basement membrane and fibronectin matrix are distinct entities in the developing mouse blastocyst. Anatomical Record, 232, 141–149.
Assou, S., Anahory, T., Pantesco, V., et al. (2006). The human cumulus–oocyte complex gene-expression profile. Human Reproduction, 21, 1705–1719.
Doyle, L. A., Yang, W., Abruzzo, L. V., et al. (1998). A multidrug resistance transporter from human MCF-7 breast cancer cells. Proceedings of the National Academy of Sciences of the United States of America, 95, 15665–15670.
Bhattacharya, B., Miura, T., Brandenberger, R., et al. (2004). Gene expression in human embryonic stem cell lines: unique molecular signature. Blood, 103, 2956–2964.
Chan, E. M., Ratanasirintrawoot, S., Park, I. H., et al. (2009). Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nature Biotechnology, 27, 1033–1037.
Mao, Q. (2008). BCRP/ABCG2 in the placenta: expression, function and regulation. Pharmaceutical Research, 25, 1244–1255.
Kolwankar, D., Glover, D. D., Ware, J. A., & Tracy, T. S. (2005). Expression and function of ABCB1 and ABCG2 in human placental tissue. Drug Metabolism and Disposition, 33, 524–529.
Simpson, A. J., Caballero, O. L., Jungbluth, A., Chen, Y. T., & Old, L. J. (2005). Cancer/testis antigens, gametogenesis and cancer. Nature Reviews. Cancer, 5, 615–625.
Scanlan, M. J., Simpson, A. J., & Old, L. J. (2004). The cancer/testis genes: review, standardization, and commentary. Cancer Immunity, 4, 1.
Jungbluth, A. A., Silva, W. A., Jr., Iversen, K., et al. (2007). Expression of cancer-testis (CT) antigens in placenta. Cancer Immunity, 7, 15.
Gjerstorff, M. F., Kock, K., Nielsen, O., & Ditzel, H. J. (2007). MAGE-A1, GAGE and NY-ESO-1 cancer/testis antigen expression during human gonadal development. Human Reproduction, 22, 953–960.
Gjerstorff, M. F., Harkness, L., Kassem, M., et al. (2008). Distinct GAGE and MAGE-A expression during early human development indicate specific roles in lineage differentiation. Human Reproduction, 23, 2194–2201.
Sigalotti, L., Coral, S., Altomonte, M., et al. (2002). Cancer testis antigens expression in mesothelioma: role of DNA methylation and bioimmunotherapeutic implications. British Journal of Cancer, 86, 979–982.
De Smet, C., De Backer, O., Faraoni, I., Lurquin, C., Brasseur, F., & Boon, T. (1996). The activation of human gene MAGE-1 in tumor cells is correlated with genome-wide demethylation. Proceedings of the National Academy of Sciences of the United States of America, 93, 7149–7153.
Klaffky, E. J., Gonzales, I. M., & Sutherland, A. E. (2006). Trophoblast cells exhibit differential responses to laminin isoforms. Developmental Biology, 292, 277–289.
Nagata, A., Ito, M., Iwata, N., et al. (1996). G protein-coupled cholecystokinin-B/gastrin receptors are responsible for physiological cell growth of the stomach mucosa in vivo. Proceedings of the National Academy of Sciences of the United States of America, 93, 11825–11830.
Noble, P. J., Wilde, G., White, M. R., Pennington, S. R., Dockray, G. J., & Varro, A. (2003). Stimulation of gastrin-CCKB receptor promotes migration of gastric AGS cells via multiple paracrine pathways. American Journal of Physiology. Gastrointestinal and Liver Physiology, 284, G75–84.
Wroblewski, L. E., Pritchard, D. M., Carter, S., & Varro, A. (2002). Gastrin-stimulated gastric epithelial cell invasion: the role and mechanism of increased matrix metalloproteinase 9 expression. Biochemical Journal, 365, 873–879.
Bourc’his, D., Xu, G. L., Lin, C. S., Bollman, B., & Bestor, T. H. (2001). Dnmt3L and the establishment of maternal genomic imprints. Science, 294, 2536–2539.
Ooi, S. K., Qiu, C., Bernstein, E., et al. (2007). DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature, 448, 714–717.
Chedin, F., Lieber, M. R., & Hsieh, C. L. (2002). The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proceedings of the National Academy of Sciences of the United States of America, 99, 16916–16921.
Bourc’his, D., & Bestor, T. H. (2004). Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature, 431, 96–99.
Mehta, D. V., Kim, Y. S., Dixon, D., & Jetten, A. M. (2002). Characterization of the expression of the retinoid-related, testis-associated receptor (RTR) in trophoblasts. Placenta, 23, 281–287.
Janatpour, M. J., Utset, M. F., Cross, J. C., et al. (1999). A repertoire of differentially expressed transcription factors that offers insight into mechanisms of human cytotrophoblast differentiation. Developmental Genetics, 25, 146–157.
Yamada, K., Ogawa, H., Tamiya, G., et al. (2000). Genomic organization, chromosomal localization, and the complete 22 kb DNA sequence of the human GCMa/GCM1, a placenta-specific transcription factor gene. Biochemical and Biophysical Research Communications, 278, 134–139.
Rawn, S. M., & Cross, J. C. (2008). The evolution, regulation, and function of placenta-specific genes. Annual Review of Cell and Developmental Biology, 24, 159–181.
Black, S. G., Arnaud, F., Palmarini, M., & Spencer, T. E. Endogenous retroviruses in trophoblast differentiation and placental development. Am J Reprod Immunol, 64, 255–264.
Ma, G. T., Roth, M. E., Groskopf, J. C., et al. (1997). GATA-2 and GATA-3 regulate trophoblast-specific gene expression in vivo. Development, 124, 907–914.
She, H., Xiong, S., Hazra, S., & Tsukamoto, H. (2005). Adipogenic transcriptional regulation of hepatic stellate cells. Journal of Biological Chemistry, 280, 4959–4967.
Xu, R. H., Chen, X., Li, D. S., et al. (2002). BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nature Biotechnology, 20, 1261–1264.
Schulz, L. C., Ezashi, T., Das, P., Westfall, S. D., Livingston, K. A., & Roberts, R. M. (2008). Human embryonic stem cells as models for trophoblast differentiation. Placenta, 29(Suppl A), S10–16.
Assou, S., Cerecedo, D., Tondeur, S., et al. (2009). A gene expression signature shared by human mature oocytes and embryonic stem cells. BMC Genomics, 10, 10.
Haouzi, D., Dechaud, H., Assou, S., Monzo, C., de Vos, J., & Hamamah, S. (2011). Transcriptome analysis reveals dialogues between human trophectoderm and endometrial cells during the implantation period. Human Reproduction
Eisen, M. B., Spellman, P. T., Brown, P. O., & Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences of the United States of America, 95, 14863–14868.
Tusher, V. G., Tibshirani, R., & Chu, G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America, 98, 5116–5121.
Al-Shahrour, F., Diaz-Uriarte, R., & Dopazo, J. (2004). FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics, 20, 578–580.
Thomson, J. A., Itskovitz-Eldor, J., Shapiro, S. S., et al. (1998). Embryonic stem cell lines derived from human blastocysts. Science, 282, 1145–1147.
Yu, J., Vodyanik, M. A., Smuga-Otto, K., et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science, 318, 1917–1920.
Strom, S., Inzunza, J., Grinnemo, K. H., et al. (2007). Mechanical isolation of the inner cell mass is effective in derivation of new human embryonic stem cell lines. Human Reproduction, 22, 3051–3058.
Acknowledgments
We thank Tanguy Le Carrour for expert help in maintaining the Amazonia! database and Robert Feil for helpful suggestions. JDV is supported by the ANR (grant ANR-07-BLAN-0076-01), JDV and SH by the Association Française contre les Myopathies (AFM), JDV by the ANR (ANR-08-BIOT-012-02) and the Région Languedoc-Roussillon (Chercheur d’Avenir 09–13198 01).
Conflicts of Interest
The authors declare no potential conflicts of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary Table S1
Microarray data list. List of the samples composing the microarray data panel, including GEO accession numbers and references. (XLS 44 kb)
Supplementary Table S2
Gene list 1: cell cycle analysis. (XLS 2159 kb)
Supplementary Table S3
Description of the 10 hESC lines used for comparison with the TE samples. (XLS 18 kb)
Supplementary Table S4
: Gene list 2: signatures. (XLS 2609 kb)
Supplementary Table S5
Detail of the 29 interactions identified by IPA in the TE core transcriptional circuitry. (XLS 19 kb)
Supplementary Table S6
Summary of the parameters and results of the SAM analyses. (XLS 9 kb)
Supplementary Table S7
Description of the primers used for real-time quantitative PCR (XLS 12 kb)
Supplementary Figure S1
Validation of the microarray data by RT-qPCR. (TIFF 1271 kb)
Supplementary Figure S2
Details of the GO terms that are significantly represented in the TE and hESC signatures. The statistical results were obtained using the FatiGO+ tool with default parameters. Only significant GO terms (p < 0.05) using an adjusted p-value were retained. Figure 3A is a graphical representation of these results. (TIFF 2187 kb)
Supplementary Figure S3
ALPPL2 splicing variants show different expression levels in TE, placenta and other tissues. (A) Expression profiling of different ALPPL2 PS illustrated by Amazonia! in all the panel samples. (B) Expected genomic positions of different ALPPL2 PS along the human ALPPL2 sequence (according to Affymetrix, data obtained on the web site http://genome.ucsc.edu/). (TIFF 7523 kb)
Supplementary Figure S4
Original IPA results showing a network of TFs that are over-expressed in TE and placenta. The interactions are numbered and the references can be found in Supplementary Table S5. (TIFF 4247 kb)
Supplementary Figure S5
Expression of the genes discussed in this study in the 181 samples of the panel. (PPT 8294 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Bai, Q., Assou, S., Haouzi, D. et al. Dissecting the First Transcriptional Divergence During Human Embryonic Development. Stem Cell Rev and Rep 8, 150–162 (2012). https://doi.org/10.1007/s12015-011-9301-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12015-011-9301-3