Abstract
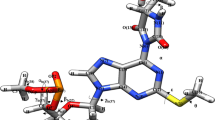
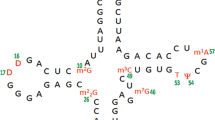
Conformational preferences of hypermodified nucleoside 5-taurinomethyluridine 5′-monophoshate ‘p-τm5U’ (–CH2–NH2 +–CH2–CH2–SO3 −) have been investigated using semi-empirical RM1 method. Automated geometry optimization using ab initio molecular orbital HF-SCF (6-31G**) and DFT (B3LYP/6-31G**) calculations have also been made to compare the salient features. The RM1 preferred most stable conformation of ‘p-τm5U’ has been stabilized by hydrogen bonding interactions between O(11a)…HN(8), O1P(34)…HN(8), and O1P(34)…HC(10). Another conformational study of 5-taurinomethyluridine side chain has also been performed in context of anticodon loop bases of E. coli tRNALeu. The atom O(11a) of τm5U(34) side chain interacts with adenosine (A35) as well as ribose–phosphate backbone which might provide structural stability to the anticodon loop. The glycosyl torsion angle of τm5U retains ‘anti’-conformation. The solvent accessible surface area calculations revealed the role of τm5U in tRNALeu anticodon loop. MD simulation results are found in agreement with RM1 preferred stable structure. The MEPs calculations of τm5U(34):G3 model show unique potential tunnels between the hydrogen bond donor and acceptor atoms as compared to τm5U(34):A3 model. Thus, these results could pave the way to understand the role of τm5U(34) to recognize UUG/UUA codons at atomic level in the mitochondrial disease, MELAS.









Similar content being viewed by others
References
Limbach, P. A., Crain, P. F., & McCloskey, J. A. (1994). Summary: The modified nucleosides of RNA. Nucleic Acids Research, 22, 2183–2196.
Curran, J. F. (1998). Modified nucleosides in translation. In H. Grosjean & R. Renne (Eds.), Modification and editing of RNA (pp. 493–516). Washington, DC: ASM Press.
Suzuki, T. (2005). Biosynthesis and function of tRNA wobble modifications. Topics in Current Genetics, 12, 23–69.
Juhling, F., Morl, M., Hartmann, R. K., Sprinzl, M., Stadler, P. F., & Putz, J. (2009). tRNAdb 2009: Compilation of tRNA sequences and tRNA genes. Nucleic Acids Research, 37, D159–D162.
Bjork, G. R. (1995). Biosynthesis and function of modified nucleosides. In D. R. Soll & U. L. RajBhandary (Eds.), tRNA: Structure, biosynthesis and function (pp. 165–205). Washington, DC: ASM Press.
Yokoyama, S., & Nishimura, S. (1995). Modified nucleosides and codon recognition. In D. R. Soll & U. L. RajBhandary (Eds.), tRNA: Structure, biosynthesis and function (pp. 207–224). Washington, DC: ASM Press.
Suzuki, T., Suzukim, T., Wada, T., Saigo, K., & Watanabe, K. (2001). Novel taurine-containing uridine derivatives and mitochondrial human diseases. Nucleic Acids Research, 1, 257–258.
Suzuki, T., Suzuki, T., Wada, T., Saigo, K., & Watanabe, K. (2002). Taurine as a constituent of mitochondrial tRNAs: New insights into the functions of taurine and human mitochondrial diseases. The EMBO Journal, 21, 6581–6589.
Yasukawa, T., Suzuki, T., Suzuki, T., Ueda, T., Ohta, S., & Watanabe, K. (2000). Modification defect at anticodon wobble nucleotide of mitochondrial tRNAsLeu(UUR) with pathogenic mutations of mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes. The Journal Biological Chemistry, 275, 4251–4257.
Cortijo, V., Sanz, M. E., Lopez, J. C., & Alonso, J. L. (2009). Conformational study of taurine in the gas phase. The Journal of Physical Chemistry A, 113, 14681–14683.
Sturman, J. A. (1993). Taurine in development. Physical Review, 73, 119–147.
Huxtable, R. J. (1992). Physiological actions of taurine. Physical Review, 72, 101–163.
Mankovskaya, I. N., Serebrovskaya, T. V., Swanson, R. J., Vavilova, G. L., & Kharlamova, O. N. (2000). Mechanisms of taurine antihypoxic and antioxidant action. High Altitude Medicine & Biology, 1, 105–110.
Pokhrel, P. K., & Lau-Cam, C. A. (2000). Protection by taurine and structurally related sulfur-containing compounds against erythrocyte membrane damage by hydrogen peroxide. Advances in Experimental Medicine and Biology, 483, 411–429.
Del Olmo, N., Galarreta, M., Bustamante, J., Del Rio, R. M., & Solis, J. M. (2000). Taurine-induced synaptic potentiation: Role of calcium and interaction with LTP. Neuropharmacology, 39, 40–54.
Bureau, M. H., & Olsen, R. W. (1991). Taurine acts on a subclass of GABAA receptors in mammalian brain in vitro. European Journal of Pharmacology, 207, 9–16.
Kirino, Y., Yasukawa, T., Ohta, S., Akira, S., Ishihara, K., Watanabe, K., & Suzuki, T. (2004). Codon-specific translational defect caused by a wobble modification deficiency in mutant tRNA from a human mitochondrial disease. Proceedings of National Academy of Sciences USA, 101, 15070–15075.
Goto, Y., Nonaka, I., & Horai, S. (1990). A mutation in the tRNA(Leu)(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature, 348, 651–653.
Goto, Y., Nonaka, I., & Horai, S. (1991). A new mtDNA mutation associated with mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes (MELAS). Biochimica et Biophysica Acta, 1097, 238–240.
Kobayashi, Y., Momoi, M. Y., Tominaga, K., Momoi, T., Nihei, K., Yanagisawa, M., et al. (1990). A point mutation in the mitochondrial tRNA(Leu)(UUR) gene in MELAS (mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes). Biochemical and Biophysical Research Communications, 173, 816–822.
Hess, J. F., Parisi, M. A., Bennett, J. L., & Clayton, D. A. (1991). Impairment of mitochondrial transcription termination by a point mutation associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature, 351, 236–239.
Flierl, A., Reichmann, H., & Seibel, P. (1997). Pathophysiology of the MELAS 3243 transition mutation. The Journal Biological Chemistry, 272, 27189–27196.
Chomyn, A., Enriquez, J. A., Micol, V., Fernandez-Silva, P., & Attardi, G. (2000). The mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episode syndrome-associated human mitochondrial tRNALeu(UUR) mutation causes aminoacylation deficiency and concomitant reduced association of mRNA with ribosomes. The Journal Biological Chemistry, 275, 19198–19209.
Jacobs, H. T. (2003). Disorders of mitochondrial protein synthesis. Human Molecular Genetics, 12, R293–R301.
Florentz, C., Sohm, B., Tryoen-Toth, P., Putz, J., & Sissler, M. (2003). Human mitochondrial tRNAs in health and disease. Cellular and Molecular Life Sciences, 60, 1356–1375.
Wittenhagen, L. M., & Kelley, S. O. (2002). Dimerization of a pathogenic human mitochondrial tRNA. Nature Structural & Molecular Biology, 9, 586–590.
Kurata, S., Weixlbaumer, A., Ohtsuki, T., Shimazaki, T., Wada, T., Kirino, Y., et al. (2008). Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U.G wobble pairing during decoding. The Journal Biological Chemistry, 283, 18801–18811.
Rocha, G. B., Freire, R. O., Simas, A. M., & Stewart, J. J. P. (2006). RM1: A reparameterization of AM1 for H, C, N, O, P, P, F, Cl, Br and I. Journal of Computational Chemistry, 27, 1101–1111.
Holbrook, S. R., Sussman, J. L., Warrant, R. W., & Kim, S. H. (1978). Crystal structure of yeast phenylalanine transfer RNA II. Structural feature and functional implications. Journal of Molecular Biology, 123, 631–660.
Sonavane, U. B., Sonawane, K. D., & Tewari, R. (2002). Conformational preferences of base substituent in hypermodified nucleotide queuosine 5′ monophosphate pQ and protonated variant pQH+. Journal of Biomolecular Structure & Dynamics, 20, 473–485.
Kennard, O. (1980–1981). In: R.C. Weast & M.J. Astle (Eds.), CRC handbook of chemistry and physics, 61st ed. (pp. F208–F211). Boca Raton, FL: CRC Press.
Ferreira, D. C., Machado, A. E. H., Tiago, F. S., Madurro, J. M., Madurro, A. G. B., & Odonirio, A. (2012). Molecular modeling study on the possible polymers formed during the electropolymerization of 3-hydroxyphenylacetic acid. Journal of Molecular Graphics and Modelling, 34, 18–27.
Labidi, N. S. (2012). Comparative study of kinetics isomerization of substituted polyacetylene (Cl, F, Br and I): Semi empirical RM1 study. Journal of Statistical Computational and Simulation. doi:10.1016/j.jscs.2012.01.008.
Pol-Fachin, L., Fraga, C. A. M., Barreiro, E. J., & Verli, H. (2010). A characterization of the conformational ensemble from bioactive N-acylhydrazone derivatives. Journal of Molecular Graphics and Modelling, 28, 446–454.
Kerber, V. A., Passos, C. S., Verli, H., Fett-Neto, A. G., Quirion, J. P., & Henriques, A. T. (2008). Psychollatine, a glucosidic monoterpene indole alkaloid from Psychotria umbellate. Journal of Natural Products, 71, 697–700.
Goncalves, A. S., Franca, T. C. C., Figueroa-Villar, J. D., & Pascutti, P. G. (2010). Conformational analysis of toxogonine, TMB-4 and HI-6 using PM6 and RM1 methods. Journal of Brazilian Chemical Society, 21, 179–184.
Anisimov, V. M., & Cavasotto, C. N. (2011). Hydration free energies using semiempirical quantum mechanical Hamiltonians and a continuum solvent model with multiple atomic-type parameters. The Journal of Physical Chemistry B, 115, 7896–7905.
Bavi, R. S., Kamble, A. D., Kumbhar, N. M., Kumbhar, B. V., & Sonawane, K. D. (2011). Conformational preferences of modified nucleoside N2-methylguanosine (m2G) and its derivative N2, N2-dimethylguanosine (m2G) occur at 26th position (Hinge Region) in tRNA. Cell Biochemistry and Biophysics, 61, 507–521.
Kumbhar, N. M., & Sonawane, K. D. (2011). Iso-energetic multiple conformations of hypermodified nucleic acid base wybutine (yW) which occur at 37th position in anticodon loop of tRNAPhe. Journal of Molecular Graphics and Modelling, 29, 935–946.
Kumbhar, N. M., Kumbhar, B. V., & Sonawane, K. D. (2012). Structural significance of hypermodified nucleic acid base hydroxywybutine (OHyW) which occur at 37th position in the anticodon loop of tRNAPhe. Journal of Molecular Graphics and Modelling, 38, 174–185.
Hehre, J., Radom, W. L., Schleyer, P. V. R., & Pople, J. A. (1986). Ab initio molecular orbital theory. New York: Wiley.
Slater, J. C. (1951). A simplification of the Hartree–Fock method. Physical Review, 81, 385–390.
Becke, A. D. (1993). Density functional thermochemistry. III. The role of exact exchange. The Journal of Chemical Physics, 98, 5648–5652.
Francl, M. M., Pietro, W. J., Hehre, W. J., Binkley, J. S., Gordon, M. S., Defrees, D. J., & Pople, J. A. (1982). Self-consistent molecular orbital methods. XXIII. A polarization-type basis set for second-row elements. The Journal of Chemical Physics, 77, 3654–3665.
Sonawane, K. D., & Tewari, R. (2008). Conformational preferences of hypermodified nucleoside lysidine (k2C) occuring at wobble position in anticodon loop of tRNAIle. Nucleoside Nucleotides and Nucleic acids., 27, 1158–1174.
Kumbhar, B. V., Kamble, A. D., & Sonawane, K. D. (2013). Conformational preferences of modified nucleoside N(4)-acetylcytidine, ac4C occur at “Wobble” 34th position in the anticodon loop of tRNA. Cell Biochemistry and Biophysics, 66, 797–816.
Tewari, R. (1987). Theoretical studies on conformational preferences of modified nucleic acid base N6-(N-threonylcarbonyl) adenine. Indian Journal of Biochemistry & Biophysics, 24, 170–176.
Tewari, R. (1987). Theoretical studies on conformational preferences of modified nucleic acid base N6-(N-glycylcarbonyl) adenine. International Journal of Quantum Chemistry, 31, 611–624.
Tewari, R. (1988). Conformational preferences of modified nucleic acid bases N6-(∆2-isopentenyl) adenine and 2-methylthio-N6-(∆2-isopentenyl) adenine by the quantum chemical PCILO calculations. International Journal of Quantum Chemistry, 34, 133–142.
Sonawane, K. D., Sonavane, U. B., & Tewari, R. (2002). Conformational preferences of anticodon 3′-adjacent hypermodified nucleic acid base cis- or trans-zeatin and its 2-methylthio derivatives cis- or trans-ms2zeatin. Journal of Biomolecular Structure & Dynamics, 19, 637–648.
Sonawane, K. D., Sonavane, U. B., & Tewari, R. (2000). Conformational flipping of the N(6) substituent in diprotonated N6-(N-glycylcarbonyl) adenines: The role of N(6) H in purine ring protonated ureido adenines. International Journal of Quantum Chemistry, 78, 398–405.
Sonavane, U. B., Sonawane, K. D., Morin, A., Grosjean, H., & Tewari, R. (1999). N(7)-protonation induced conformational flipping in hypermodified nucleic acid bases N6-(N-threonylcarbonyl) adenine and its 2-methylthio- or N(6)-methyl-derivatives. International Journal of Quantum Chemistry, 75, 223–229.
SYBYL 7.3. (2006) Tripos International, South Hanley Rd., St. Louis, MO, USA.
Cornell, W. D., Cieplak, P., Bayly, C. I., Gould, I. R., Merz, K. M, Jr, Ferguson, D. M., et al. (1995). A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. Journal of the American Chemical Society, 117, 5179–5197.
Takai, K., & Yokoyama, S. (2003). Roles of 5-substituents of tRNA wobble uridines in the recognition of purine-ending codons. Nucleic Acids Research, 31, 6383–6391.
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., & Ferrin, T. E. (2004). UCSF Chimera—A visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25, 1605–1612.
Bavi, R. S., Sambhare, S. B., & Sonawane, K. D. (2013). MD simulation studies to investigate iso-energetic conformational behavior of modified nucleosides m2G and m 22 G present in the tRNA. Computational and Structural Biotechnology Journal, 5, e201302015.
Murphy, F. V., Ramakrishnan, V., Malkiewicz, A., & Agris, P. F. (2004). The role of modifications in codon discrimination by tRNALys UUU. Nature Structural & Molecular Biology, 11, 1186–1191.
Crick, F. H. C. (1966). Codon-anticodon pairing: The wobble hypothesis. Journal of Molecular Biology, 19, 548–555.
Otero-Navas, I., & Seminario, J. M. (2012). Molecular electrostatic potential of DNA base-base pairing and mispairing. Journal of Molecular Modelling., 18, 91–101.
Acknowledgments
KDS is gratefully acknowledged to University Grants Commission (UGC), New Delhi for financial support under the major research Project, vide UGC Letter No. F. 40-204/2011 (SR) dated June 29, 2011. ASK, BVK, and RSB are thankful to UGC for providing project fellowship.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kamble, A.S., Kumbhar, B.V., Sambhare, S.B. et al. Conformational Preferences of Modified Nucleoside 5-Taurinomethyluridine, τm5U Occur at ‘wobble’ 34th Position in the Anticodon Loop of tRNA. Cell Biochem Biophys 71, 1589–1603 (2015). https://doi.org/10.1007/s12013-014-0382-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12013-014-0382-x