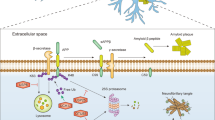
Abstract
Ubiquitin proteasome system (UPS) determines the timing and extent of protein turnover in cells, and it is one of the most strictly controlled cellular mechanisms. Lack of proper control over UPS is attributed to both cancer and to neurodegenerative diseases, yet in different context and direction. Cancerous cells have altered cellular metabolisms, uncontrolled cellular division, and increased proteasome activity. The specialized function prevent neurons from undergoing cellular division but allow them to extend an axon over long distances, establish connections, and to form stable neuronal circuitries. Neurons heavily depend on the proper function of the proteasome and the UPS for their proper function. Reduction of UPS function in vulnerable neurons results in protein aggregation, increased ER stress, and cell death. Identification of compounds that selectively block proteasome function in distinct set of malignancies added momentum to drug discovery efforts, and deubiquitinases (DUBs) gained much attention. This review will focus on ubiquitin carboxy-terminal hydrolase L1 (UCHL1), a DUB that is attributed to both cancer and neurodegeneration. The potential of developing effective treatment strategies for two major health problems by controlling the function of UPS opens up new avenues for innovative approaches and therapeutic interventions.

Similar content being viewed by others
References
Ciechanover, A., Heller, H., Elias, S., et al. (1980). ATP-dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proceedings of the National Academy of Sciences of the United States of America, 77(3), 1365–1368.
Hershko, A., Ciechanover, A., Heller, H., et al. (1980). Proposed role of ATP in protein breakdown: conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proceedings of the National Academy of Sciences of the United States of America, 77(4), 1783–1786.
Varshavsky, A. (2012). The ubiquitin system, an immense realm. Annual Review of Biochemistry, 81, 167–176.
Xu, G., & Jaffrey, S. R. (2011). The new landscape of protein ubiquitination. Nature Biotechnology, 29(12), 1098–1100.
Neutzner, M., & Neutzner, A. (2012). Enzymes of ubiquitination and deubiquitination. Essays in Biochemistry, 52, 37–50.
Liu, J., & Nussinov, R. (2012). The role of allostery in the ubiquitin-proteasome system. Critical Reviews in Biochemistry and Molecular Biology. doi:10.3109/10409238.2012.742856.
Komander, D., & Rape, M. (2012). The ubiquitin code. Annual Review of Biochemistry, 81, 203–229.
Chhangani, D., Joshi, A. P., & Mishra, A. (2012). E3 ubiquitin ligases in protein quality control mechanism. Molecular Neurobiology, 45(3), 571–585.
Lee, M. J., Lee, B. H., Hanna, J., et al. (2011). Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Molecular and Cellular Proteomics, 10(5), R110003871.
Nijman, S. M., Luna-Vargas, M. P., Velds, A., et al. (2005). A genomic and functional inventory of deubiquitinating enzymes. Cell, 123(5), 773–786.
Sowa, M. E., Bennett, E. J., Gygi, S. P., et al. (2009). Defining the human deubiquitinating enzyme interaction landscape. Cell, 138(2), 389–403.
Todi, S. V., & Paulson, H. L. (2011). Balancing act: Deubiquitinating enzymes in the nervous system. Trends in Neurosciences, 34(7), 370–382.
Day, I. N., & Thompson, R. J. (2010). UCHL1 (PGP 9.5): Neuronal biomarker and ubiquitin system protein. Progress in Neurobiology, 90(3), 327–362.
Day, I. N., & Thompson, R. J. (1987). Molecular cloning of cDNA coding for human PGP 9.5 protein. A novel cytoplasmic marker for neurones and neuroendocrine cells. FEBS Letters, 210(2), 157–160.
Mermerian, A. H., Case, A., Stein, R. L., et al. (2007). Structure-activity relationship, kinetic mechanism, and selectivity for a new class of ubiquitin C-terminal hydrolase-L1 (UCH-L1) inhibitors. Bioorganic and Medicinal Chemistry Letters, 17(13), 3729–3732.
Hirayama, K., Aoki, S., Nishikawa, K., et al. (2007). Identification of novel chemical inhibitors for ubiquitin C-terminal hydrolase-L3 by virtual screening. Bioorganic and Medicinal Chemistry, 15(21), 6810–6818.
Liu, Y., Fallon, L., Lashuel, H. A., et al. (2002). The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson’s disease susceptibility. Cell, 111(2), 209–218.
Luchansky, S. J., Lansbury, P. T, Jr, & Stein, R. L. (2006). Substrate recognition and catalysis by UCH-L1. Biochemistry, 45(49), 14717–14725.
Meray, R. K., & Lansbury, P. T, Jr. (2007). Reversible monoubiquitination regulates the Parkinson disease-associated ubiquitin hydrolase UCH-L1. Journal of Biological Chemistry, 282(14), 10567–10575.
Osaka, H., Wang, Y. L., Takada, K., et al. (2003). Ubiquitin carboxy-terminal hydrolase L1 binds to and stabilizes monoubiquitin in neuron. Human Molecular Genetics, 12(16), 1945–1958.
Saigoh, K., Wang, Y. L., Suh, J. G., et al. (1999). Intragenic deletion in the gene encoding ubiquitin carboxy-terminal hydrolase in gad mice. Nature Genetics, 23(1), 47–51.
Calzada, B., Naves, F. J., Del Valle, M. E., et al. (1994). Distribution of protein gene product 9.5 (PGP 9.5) immunoreactivity in the dorsal root ganglia of adult rat. Annals of Anatomy, 176(5), 437–441.
Schofield, J. N., Day, I. N., Thompson, R. J., et al. (1995). PGP9.5, a ubiquitin C-terminal hydrolase; pattern of mRNA and protein expression during neural development in the mouse. Brain Research. Developmental Brain Research, 85(2), 229–238.
Weis, J., Katona, I., Muller-Newen, G., et al. (2011). Small-fiber neuropathy in patients with ALS. Neurology, 76(23), 2024–2029.
Kabuta, T., Setsuie, R., Mitsui, T., et al. (2008). Aberrant molecular properties shared by familial Parkinson’s disease-associated mutant UCH-L1 and carbonyl-modified UCH-L1. Human Molecular Genetics, 17(10), 1482–1496.
Proctor, C. J., Tangeman, P. J., & Ardley, H. C. (2010). Modelling the role of UCH-L1 on protein aggregation in age-related neurodegeneration. PLoS ONE, 5(10), e13175.
Setsuie, R., & Wada, K. (2007). The functions of UCH-L1 and its relation to neurodegenerative diseases. Neurochemistry International, 51(2–4), 105–111.
Zhong, J., Zhao, M., Ma, Y., et al. (2012). UCHL1 acts as a colorectal cancer oncogene via activation of the beta-catenin/TCF pathway through its deubiquitinating activity. International Journal of Molecular Medicine, 30(2), 430–436.
Trifa, F., Karray-Chouayekh, S., Jmaa, Z. B., et al. (2013). Frequent CpG methylation of ubiquitin carboxyl-terminal hydrolase 1 (UCHL1) in sporadic and hereditary Tunisian breast cancer patients: clinical significance. Medical Oncology, 30(1), 418.
Xiang, T., Li, L., Yin, X., et al. (2012). The ubiquitin peptidase UCHL1 induces G0/G1 cell cycle arrest and apoptosis through stabilizing p53 and is frequently silenced in breast cancer. PLoS ONE, 7(1), e29783.
Ummanni, R., Jost, E., Braig, M., et al. (2011). Ubiquitin carboxyl-terminal hydrolase 1 (UCHL1) is a potential tumour suppressor in prostate cancer and is frequently silenced by promoter methylation. Molecular Cancer, 10, 129.
Rao, Z., & Ding, Y. (2012). Ubiquitin pathway and ovarian cancer. Current Oncology, 19(6), 324–328.
Carolan, B. J., Heguy, A., Harvey, B. G., et al. (2006). Up-regulation of expression of the ubiquitin carboxyl-terminal hydrolase L1 gene in human airway epithelium of cigarette smokers. Cancer Research, 66(22), 10729–10740.
D’Arcy, P., & Linder, S. (2012). Proteasome deubiquitinases as novel targets for cancer therapy. International Journal of Biochemistry and Cell Biology, 44(11), 1729–1738.
Poulsen, J. W., Madsen, C. T., Young, C., et al. (2012). Comprehensive profiling of proteome changes upon sequential deletion of deubiquitylating enzymes. Journal of Proteomics, 75(13), 3886–3897.
Driscoll, J. J., Minter, A., Driscoll, D. A., et al. (2011). The ubiquitin+proteasome protein degradation pathway as a therapeutic strategy in the treatment of solid tumor malignancies. Anti-Cancer Agents in Medicinal Chemistry, 11(2), 242–246.
Cvek, B., & Dvorak, Z. (2011). The ubiquitin-proteasome system (UPS) and the mechanism of action of bortezomib. Current Pharmaceutical Design, 17(15), 1483–1499.
Rzymski, T., Petry, A., Kracun, D., et al. (2012). The unfolded protein response controls induction and activation of ADAM17/TACE by severe hypoxia and ER stress. Oncogene, 31(31), 3621–3634.
McLaughlin, M., & Vandenbroeck, K. (2011). The endoplasmic reticulum protein folding factory and its chaperones: New targets for drug discovery? British Journal of Pharmacology, 162(2), 328–345.
Hong, Y., Ho, K. S., Eu, K. W., et al. (2007). A susceptibility gene set for early onset colorectal cancer that integrates diverse signaling pathways: implication for tumorigenesis. Clinical Cancer Research, 13(4), 1107–1114.
Mandelker, D. L., Yamashita, K., Tokumaru, Y., et al. (2005). PGP9.5 promoter methylation is an independent prognostic factor for esophageal squamous cell carcinoma. Cancer Research, 65(11), 4963–4968.
Sato, F., & Meltzer, S. J. (2006). CpG island hypermethylation in progression of esophageal and gastric cancer. Cancer, 106(3), 483–493.
Lee, Y. M., Lee, J. Y., Kim, M. J., et al. (2006). Hypomethylation of the protein gene product 9.5 promoter region in gallbladder cancer and its relationship with clinicopathological features. Cancer Science, 97(11), 1205–1210.
Mizukami, H., Shirahata, A., Goto, T., et al. (2008). PGP9.5 methylation as a marker for metastatic colorectal cancer. Anticancer Research, 28(5A), 2697–2700.
Frisan, T., Coppotelli, G., Dryselius, R., et al. (2012). Ubiquitin C-terminal hydrolase-L1 interacts with adhesion complexes and promotes cell migration, survival, and anchorage independent growth. FASEB Journal, 26(12), 5060–5070.
Kim, H. J., Kim, Y. M., Lim, S., et al. (2009). Ubiquitin C-terminal hydrolase-L1 is a key regulator of tumor cell invasion and metastasis. Oncogene, 28(1), 117–127.
Bheda, A., Yue, W., Gullapalli, A., et al. (2009). Positive reciprocal regulation of ubiquitin C-terminal hydrolase L1 and beta-catenin/TCF signaling. PLoS ONE, 4(6), e5955.
Bheda, A., Shackelford, J., & Pagano, J. S. (2009). Expression and functional studies of ubiquitin C-terminal hydrolase L1 regulated genes. PLoS ONE, 4(8), e6764.
Gestwicki, J. E., & Garza, D. (2012). Protein quality control in neurodegenerative disease. Progress in Molecular Biology and Translational Science, 107, 327–353.
Miller, R. J., & Wilson, S. M. (2003). Neurological disease: UPS stops delivering! Trends in Pharmacological Sciences, 24(1), 18–23.
Goedert, M., Spillantini, M. G., Del Tredici, K., et al. (2012). 100 years of Lewy pathology. Nature Reviews Neurology, 9(1), 13–24.
Spillantini, M. G., Crowther, R. A., Jakes, R., et al. (1998). alpha-Synuclein in filamentous inclusions of Lewy bodies from Parkinson’s disease and dementia with lewy bodies. Proceedings of the National Academy of Sciences of the United States of America, 95(11), 6469–6473.
Neumann, M., Sampathu, D. M., Kwong, L. K., et al. (2006). Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science, 314(5796), 130–133.
Prudencio, M., Durazo, A., Whitelegge, J. P., et al. (2010). An examination of wild-type SOD1 in modulating the toxicity and aggregation of ALS-associated mutant SOD1. Human Molecular Genetics, 19(24), 4774–4789.
Giliberto, L., d’Abramo, C., Acker, C. M., et al. (2010). Transgenic expression of the amyloid-beta precursor protein-intracellular domain does not induce Alzheimer’s disease-like traits in vivo. PLoS ONE, 5(7), e11609.
Carvalho, A. N., Marques, C., Rodrigues, E., et al. (2012). Ubiquitin-Proteasome System Impairment and MPTP-Induced Oxidative Stress in the Brain of C57BL/6 Wild-type and GSTP Knockout Mice. Molecular Neurobiology. doi:10.1007/s12035-012-8368-4.
Komatsu, M., Waguri, S., Chiba, T., et al. (2006). Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature, 441(7095), 880–884.
Anderson, C., Crimmins, S., Wilson, J. A., et al. (2005). Loss of Usp14 results in reduced levels of ubiquitin in ataxia mice. Journal of Neurochemistry, 95(3), 724–731.
Walters, B. J., Campbell, S. L., Chen, P. C., et al. (2008). Differential effects of Usp14 and Uch-L1 on the ubiquitin proteasome system and synaptic activity. Molecular and Cellular Neuroscience, 39(4), 539–548.
Crimmins, S., Jin, Y., Wheeler, C., et al. (2006). Transgenic rescue of ataxia mice with neuronal-specific expression of ubiquitin-specific protease 14. Journal of Neuroscience, 26(44), 11423–11431.
Cartier, A. E., Djakovic, S. N., Salehi, A., et al. (2009). Regulation of synaptic structure by ubiquitin C-terminal hydrolase L1. Journal of Neuroscience, 29(24), 7857–7868.
Leroy, E., Boyer, R., Auburger, G., et al. (1998). The ubiquitin pathway in Parkinson’s disease. Nature, 395(6701), 451–452.
Lowe, J., McDermott, H., Landon, M., et al. (1990). Ubiquitin carboxyl-terminal hydrolase (PGP 9.5) is selectively present in ubiquitinated inclusion bodies characteristic of human neurodegenerative diseases. Journal of Pathology, 161(2), 153–160.
Choi, J., Levey, A. I., Weintraub, S. T., et al. (2004). Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson’s and Alzheimer’s diseases. Journal of Biological Chemistry, 279(13), 13256–13264.
Barrachina, M., Castano, E., Dalfo, E., et al. (2006). Reduced ubiquitin C-terminal hydrolase-1 expression levels in dementia with Lewy bodies. Neurobiology of Diseases, 22(2), 265–273.
Lederer, C. W., Torrisi, A., Pantelidou, M., et al. (2007). Pathways and genes differentially expressed in the motor cortex of patients with sporadic amyotrophic lateral sclerosis. BMC genomics, 8, 26.
Gong, B., Cao, Z., Zheng, P., et al. (2006). Ubiquitin hydrolase Uch-L1 rescues beta-amyloid-induced decreases in synaptic function and contextual memory. Cell, 126(4), 775–788.
Hsu, S. H., Lai, M. C., Er, T. K., et al. (2010). Ubiquitin carboxyl-terminal hydrolase L1 (UCHL1) regulates the level of SMN expression through ubiquitination in primary spinal muscular atrophy fibroblasts. Clinica Chimica Acta, 411(23–24), 1920–1928.
Cottrell, B. A., Galvan, V., Banwait, S., et al. (2005). A pilot proteomic study of amyloid precursor interactors in Alzheimer’s disease. Annals of Neurology, 58(2), 277–289.
Caballero, O. L., Resto, V., Patturajan, M., et al. (2002). Interaction and colocalization of PGP9.5 with JAB1 and p27(Kip1). Oncogene, 21(19), 3003–3010.
Lakshmana, M. K., Chung, J. Y., Wickramarachchi, S., et al. (2010). A fragment of the scaffolding protein RanBP9 is increased in Alzheimer’s disease brains and strongly potentiates amyloid-beta peptide generation. FASEB Journal, 24(1), 119–127.
Zhang, M., Deng, Y., Luo, Y., et al. (2012). Control of BACE1 degradation and APP processing by ubiquitin carboxyl-terminal hydrolase L1. Journal of Neurochemistry, 120(6), 1129–1138.
Healy, D. G., Abou-Sleiman, P. M., Casas, J. P., et al. (2006). UCHL-1 is not a Parkinson’s disease susceptibility gene. Annals of Neurology, 59(4), 627–633.
Maraganore, D. M., Lesnick, T. G., Elbaz, A., et al. (2004). UCHL1 is a Parkinson’s disease susceptibility gene. Annals of Neurology, 55(4), 512–521.
Bilguvar, K., Tyagi, N. K., Ozkara, C., et al. (2013). Recessive loss of function of the neuronal ubiquitin hydrolase UCHL1 leads to early-onset progressive neurodegeneration. Proceedings of the National Academy of Sciences of the United States of America. doi:10.1073/pnas.1222732110.
Johnson, J. O., Mandrioli, J., Benatar, M., et al. (2010). Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron, 68(5), 857–864.
Hirano, M. (2008). VAPB: New genetic clues to the pathogenesis of ALS. Neurology, 70(14), 1161–1162.
Deng, H. X., Chen, W., Hong, S. T., et al. (2011). Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature, 477(7363), 211–215.
Fecto, F., Yan, J., Vemula, S. P., et al. (2011). SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Archives of Neurology, 68(11), 1440–1446.
Shukla, V., Mishra, S. K., & Pant, H. C. (2011). Oxidative stress in neurodegeneration. Advances in Pharmacological Sciences, 20115, 72634.
Rosen, D. R. (1993). Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature, 364(6435), 362.
Bosco, D. A., Morfini, G., Karabacak, N. M., et al. (2010). Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nature Neuroscience, 13(11), 1396–1403.
Castegna, A., Aksenov, M., Aksenova, M., et al. (2002). Proteomic identification of oxidatively modified proteins in Alzheimer’s disease brain. Part I: Creatine kinase BB, glutamine synthase, and ubiquitin carboxy-terminal hydrolase L-1. Free Radical Biology and Medicine, 33(4), 562–571.
Zhou, Y., Gu, G., Goodlett, D. R., et al. (2004). Analysis of alpha-synuclein-associated proteins by quantitative proteomics. Journal of Biological Chemistry, 279(37), 39155–39164.
Woodman, B., Butler, R., Landles, C., et al. (2007). The Hdh(Q150/Q150) knock-in mouse model of HD and the R6/2 exon 1 model develop comparable and widespread molecular phenotypes. Brain Research Bulletin, 72(2–3), 83–97.
Li, L., Tao, Q., Jin, H., et al. (2010). The tumor suppressor UCHL1 forms a complex with p53/MDM2/ARF to promote p53 signaling and is frequently silenced in nasopharyngeal carcinoma. Clinical Cancer Research, 16(11), 2949–2958.
Ewing, R. M., Chu, P., Elisma, F., et al. (2007). Large-scale mapping of human protein–protein interactions by mass spectrometry. Molecular Systems Biology, 3, 89.
Paudel, N., Sadagopan, S., Balasubramanian, S., et al. (2012). Kaposi’s sarcoma-associated herpesvirus latency-associated nuclear antigen and angiogenin interact with common host proteins, including annexin A2, which is essential for survival of latently infected cells. Journal of Virology, 86(3), 1589–1607.
Deribe, Y. L., Wild, P., Chandrashaker, A., et al. (2009). Regulation of epidermal growth factor receptor trafficking by lysine deacetylase HDAC6. Science Signaling, 2(102), ra84.
Naiki, T., Nagaki, M., Shidoji, Y., et al. (2002). Analysis of gene expression profile induced by hepatocyte nuclear factor 4alpha in hepatoma cells using an oligonucleotide microarray. Journal of Biological Chemistry, 277(16), 14011–14019.
Altun, M., Kramer, H. B., Willems, L. I., et al. (2011). Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chemistry and Biology, 18(11), 1401–1412.
Acknowledgments
This study was supported by Les Turner ALS Foundation, Wenske Foundation, and Brain Research Foundation Grants (P.H.O.). J.H.J. is supported by ALSA Safenowitz postdoctoral fellowship. The authors thank Dr. Baris Genc and Michael Tu for comments and critical reading of the manuscript.
Conflict of interest
None
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jara, J.H., Frank, D.D. & Özdinler, P.H. Could Dysregulation of UPS be a Common Underlying Mechanism for Cancer and Neurodegeneration? Lessons from UCHL1. Cell Biochem Biophys 67, 45–53 (2013). https://doi.org/10.1007/s12013-013-9631-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12013-013-9631-7