Abstract
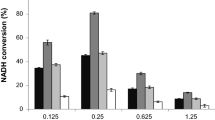
Formate dehydrogenases are critical tools for nicotinamide cofactor regeneration, but their limited catalytic efficiency (kcat/Km) is a major drawback. A formate dehydrogenase from Burkholderia stabilis 15516 (BstFDH) was the first native NADP+-dependent formate dehydrogenase reported and has the highest kcat/Km toward NADP+ (kcat/KmNADP+) compared with other FDHs that can utilize NADP+ as a hydrogen acceptor. However, the substrate and cofactor affinities of BstFDH are inferior to those of other FDHs, making its practical application difficult. Herein, we engineered recombinant BstFDH to enhance its HCOO− and NADP+ affinities. Based on sequence information analysis and homologous modeling results, I124, G146, S262, and A287 were found to affect the binding affinity for HCOO− and NADP+. By combining these mutations, we identified a BstFDH variant (G146M/A287G) that reduced KmNADP+ to 0.09 mM, with a concomitant decrease in KmHCOO−, and gave 1.6-fold higher kcat/KmNADP+ than the wild type (WT). Furthermore, BstFDH I124V/G146H/A287G, with the lowest KmHCOO− of 8.51 mM, showed a catalytic efficiency that was 2.3-fold higher than that of the wild type and a decreased KmNADP+ of 0.11 mM. These results are beneficial for improving the performance of NADP+-dependent formate dehydrogenase in the NADPH regeneration of various bioreductive reactions and provide a useful guide for engineering of the substrate and cofactor affinity of other enzymes.




Similar content being viewed by others
References
Matsuda, T., Yamanaka, R., & Nakamura, K. (2009). Recent progress in biocatalysis for asymmetric oxidation and reduction. Tetrahedron-Asymmetry, 20(5), 513–557.
Fischer, T., & Pietruszka, J. (2010). Key building blocks via enzyme mediated synthesis. Topics in Current Chemistry, 297, 1–43.
Wohlgemuth, R. (2010). Asymmetric biocatalysis with microbial enzymes and cells. Current Opinion in Microbiology, 13(3), 283–292.
Wei, P., Cui, Y. H., Zong, M. H., Xu, P., Zhou, J., & Lou, W. Y. (2017). Enzymatic characterization of a recombinant carbonyl reductase from Acetobacter sp. CCTCC M209061. Bioresour Bioprocess, 4(1), 39.
Donk, W. A. V. D., & Zhao, H. (2003). Recent developments in pyridine nucleotide regeneration. Current Opinion in Biotechnology, 14(4), 421–426.
Wichmann, R., & Vasic-Racki, D. (2005). Cofactor regeneration at the lab scale. Advances in Biochemical Engineering/Biotechnology, 92, 225–260.
Zhao, H., & Donk, W. A. V. D. (2003). Regeneration of cofactors for use in biocatalysis. Current Opinion in Biotechnology, 14(6), 583–589.
Tishkov, V. I., & Popov, V. O. (2006). Protein engineering of formate dehydrogenase. Biomolecular Engineering, 23(2–3), 89–110.
Bommarius, A. S., Schwarm, M., Stingl, K., Kottenhahn, M., Huthmacher, K., & Drauz, K. (1995). Synthesis and use of enantiometrically pure tert-leucine. Tetrahedron-Asymmetry, 6(12), 2851–2888.
Gul-Karaguler, N., Sessions, R. B., Clarke, A. R., & Holbrook, J. J. (2001). A single mutation in the NAD-specific formate dehydrogenase from Candida methylica allows the enzyme to use NADP. Biotechnology Letters, 23(4), 283–287.
Mitsuhashi, K., Yamamoto, H., & Kimoto, N. (2004). Mutants of microbacterium vaccae-derived formate dehydrogenase and uses thereof. US6830907–B2.
Andreadeli, A., Platis, D., Tishkov, V., Popov, V., & Labrou, N. E. (2008). Structure-guided alteration of coenzyme specificity of formate dehydrogenase by saturation mutagenesis to enable efficient utilization of NADP. The FEBS Journal, 275(15), 3859–3869.
Alekseeva, A. A., Fedorchuk, V. V., Zarubina, S. A., Sadykhov, E. G., Matorin, A. D., Savin, S. S., & Tishkov, V. I. (2015). Role of Ala198 in stability and coenzyme specificity of bacterial formate dehydrogenases. Acta Naturae, 7(1), 60–69.
Slusarczyk, H., Felber, S., Kula, M. R., & Pohl, M. (2003). Novel mutants of formate dehydrogenase from Candida boidinii. US20030157664–A1.
Alekseeva, A. A., Serenko, A. A., Kargov, I. S., Savin, S. S., Kleymenov, S. Y., & Tishkov, V. I. (2012). Engineering catalytic properties and thermal stability of plant formate dehydrogenase by single-point mutations. Protein Engineering, Design & Selection, 11(25), 781–788.
Kargov, I. S., Kleymenov, S. Y., Savin, S. S., Tishkov, V. I., & Alekseeva, A. A. (2015). Improvement of the soy formate dehydrogenase properties by rational design. Protein Engineering, Design & Selection, 28(6), 171–178.
Jiang, W., Lin, P., Yang, R., & Fang, B. (2016). Identification of catalysis, substrate, and coenzyme binding sites and improvement catalytic efficiency of formate dehydrogenase from Candida boidinii. Applied Microbiology and Biotechnology, 100(19), 8425–8437.
Hoelsch, K., Sührer, I., Heusel, M., & Weuster-Botz, D. (2013). Engineering of formate dehydrogenase: synergistic effect of mutations affecting cofactor specificity and chemical stability. Applied Microbiology and Biotechnology, 97(6), 2473–2481.
Hatrongjit, R., & Packdibamrung, K. (2010). A novel NADP+-dependent formate dehydrogenase from Burkholderia stabilis 15516: screening, purification and characterization. Enzyme and Microbial Technology, 46(7), 557–561.
Fogal, S., Beneventi, E., Cendron, L., & Bergantino, E. (2015). Structural basis for double cofactor specificity in a new formate dehydrogenase from the acidobacterium Granulicella mallensis MP5ACTX8. Applied Microbiology and Biotechnology, 99(22), 9541–9554.
Wu, W., Zhu, D., & Hua, L. (2009). Site-saturation mutagenesis of formate dehydrogenase from Candida bodinii creating effective NADP+-dependent FDH enzymes. Journal of Molecular Catalysis B: Enzymatic, 61(3–4), 157–116.
Guo, Q., Gakhar, L., Wickersham, K., Francis, K., Vardi-Kilshtain, A., Major, D. T., Cheatum, C. M., & Kohen, A. (2016). Structural and kinetic studies of formate dehydrogenase from Candida boidinii. Biochemistry, 55(19), 2760–2771.
Sun, Z., Salas, P. T., Siirola, E., Lonsdale, R., & Reetz, M. T. (2016). Exploring productive sequence space in directed evolution using binary patterning versus conventional mutagenesis strategies. Bioresour Bioprocess, 3(1), 44.
Cahn, J. K. B., Baumschlager, A., Brinkmann-Chen, S., & Arnold, F. H. (2015). Mutations in adenine-binding pockets enhance catalytic properties of NAD(P)H-dependent enzymes. Protein Engineering, Design & Selection, 29(1), 31–38.
You, Z. N., Chen, Q., Shi, S. C., Zheng, M. M., Pan, J., Qian, X. L., Li, C. X., & Xu, J. H. (2019). Switching cofactor dependence of 7β-hydroxysteroid dehydrogenase for cost-effective production of ursodeoxycholic acid. ACS Catalysis, 9(1), 466–473.
Lamzin, V. S., Aleshin, A. E., Strokopytov, B. V., Yukhnevich, M. G., Popov, V. O., Harutyunyan, E. H., & Wilson, K. S. (1992). Crystal structure of NAD-dependent formate dehydrogenase. European Journal of Biochemistry, 206(2), 441–452.
Torres, R. A., Schiøtt, B., & Bruice, T. C. (1999). Molecular dynamics simulations of ground and transition states for the hydride transfer from formate to NAD+ in the active site of formate dehydrogenase. Journal of the American Chemical Society, 121(36), 8164–8173.
Tishkov, V. I., & Popov, V. O. (2004). Catalytic mechanism and application of formate dehydrogenase. Biochemistry, 69(11), 1252–1267.
Alpdağştas, S., Yücel, S., Kapkaç, H. A., Liu, S., & Binay, B. (2018). Discovery of an acidic, thermostable and highly NADP+ dependent formate dehydrogenase from Lactobacillus buchneri NRRL B-30929. Biotechnology Letters, 40(7), 1135–1147.
Serov, A. E., Popova, A. S., Fedorchuk, V. V., & Tishkov, V. I. (2002). Engineering of coenzyme specificity of formate dehydrogenase from Saccharomyces cerevisiae. The Biochemical Journal, 367(3), 841–847.
Benjamin Guy, D., Ayhan, C., Gideon John, D., & Karen Mary, R. (2010). Novel enzyme. WO2010041055–A2.
Ding, H. T., Liu, D. F., Li, Z. L., Du, Y. Q., Xu, X. H., & Zhao, Y. H. (2011). Characterization of a thermally stable and organic solvent–adaptative NAD+-dependent formate dehydrogenase from Bacillus sp. F1. Journal of Applied Microbiology, 111(5), 1075–1085.
Andreadeli, A., Flemetakis, E., Axarli, I., Dimou, M., Udvardi, M. K., Katinakis, P., & Labrou, N. E. (2009). Cloning and characterization of Lotus japonicas formate dehydrogenase: A possible correlation with hypoxia. Biochimica et Biophysica Acta, 1794(6), 976–984.
Acknowledgments
We thank Simon Partridge, Ph.D., from Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (Nos. 21536004, 21871085, and 21878085), the National Key Research and Development Program of China (No. 2018YFC1706200), and the Fundamental Research Funds for the Central Universities (No. 22221818014).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
ESM 1
(PDF 946 kb)
Rights and permissions
About this article
Cite this article
Jiang, HW., Chen, Q., Pan, J. et al. Rational Engineering of Formate Dehydrogenase Substrate/Cofactor Affinity for Better Performance in NADPH Regeneration. Appl Biochem Biotechnol 192, 530–543 (2020). https://doi.org/10.1007/s12010-020-03317-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-020-03317-7