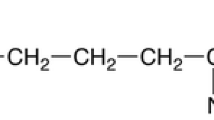Abstract
Here we first improved the ε-PL productivity in five species of wild-type strains in Streptomyces (Streptomyces padanus, Streptomyces griseofuscus, Streptomyces graminearus, Streptomyces hygroscopicus, and Streptomyces albulus) by genome shuffling. Then all the shuffled strains were suffered from an interspecific hybridization through stochastic protoplast fusion. One hybrid designated FEEL-1 was selected by morphology and spore color with ε-PL production of 1.12 g/L in shake flask, about 2.75-fold higher than that in wild types. The ε-PL production of FEEL-1 was then obtained as 24.5 g/L in fed-batch fermentation, which was 63–81 % higher than those in shuffled strains. Random amplified polymorphic DNA revealed that FEEL-1 was probably hybridized from S. padanus, S. griseofuscus, and S. albulus. Activities of several enzymes in FEEL-1 (hexokinase, phosphoenolpyruvate carboxylase, aspartokinase, and citrate synthase) were more active than those in shuffled strains, which was a possible reason for the enhancement of ε-PL production. This research highlights the importance of genome shuffling along with interspecific hybridization as a new breeding strategy for improving phenotype of industrial strains.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
ε-poly-l-lysine (ε-PL) is a homo-poly-amino acid where L-lysine monomers are linked by peptide bonds between the carboxyl and the epsilon-amino groups. Because it is water soluble, biodegradable, edible, and non-toxic toward humans and environment, ε-PL and its derivatives have been of interest for a broad range of industrial applications such as food preservatives, emulsifying agent, dietary agent, biodegradable fibers, highly water-absorbable hydrogels, drug carriers, etc. [1, 2]. Therefore, the commercial production of ε-PL is considered highly promising.
In our previous work, five species of ε-PL-producing strains (Streptomyces padanus, Streptomyces griseofuscus, Streptomyces graminearus, Streptomyces hygroscopicus, and Streptomyces albulus) were screened from soils. However, the productivity of ε-PL in these wild-type strains was low, only 0.38–0.49 g/L in shake flask and 7.0–8.0 g/L in fed-batch fermentation [3]. Comparing with other currently reported strains, this level gap was very big. For example, a mutant of S. albulus no. 410 produced ε-PL as many as 48.3 g/L in fed-batch fermentation [4], which is still the highest yield at present; Zhang et al. [5] described a mutant named Kitasatospora sp. MY 5–36 which seemed to be able to produce up to 22 g/L in liquid culture and 34 g/L with immobilized cells; Chen et al. claimed a high production of ε-PL as 35.1 g/L in strain M-Z18 using glucose and glycerol as a mixed carbon source [6]. Therefore, mass of significative work should focus on strain breeding to meet industrial needs. Although metabolic and protein engineering are common strategies for the manipulation of organisms to synthesize high-value compounds effectively, they are suitable for strains whose details of biosynthetic pathway and regulatory mechanism are characterized. But for our wild-type strains, the genetic backgrounds remain unclear and it is impractical to obtain mutants with remarkably improved ε-PL production in short period by these rational methods.
Fortunately, genome shuffling has recently been a powerful means for rapid breeding of improved organisms [7–12] which can be applicable to the improvement of most production pathways and other complex phenotypes in uncharacterized bacteria. We have reported that genome shuffling enhanced ε-PL production of S. graminearus by improving glucose tolerance [9] and proved the efficiency of this approach. In this research, we adopted genome shuffling to increase ε-PL productivity of the five wild-type strains by glucose (GLU) tolerance, sulfaguanidine (SG) tolerance, and succinic acid (SA) as sole carbon source, respectively. Indeed, the shuffled strains showed improvements in ε-PL production; however, titers remained low (13.5–16.9 g/L). We postulated that lack of coordination for integral metabolism occurred in shuffled strains because contributions to improvement of ε-PL production from higher activity of single enzyme were limited. To overcome this constraint, we carried out an interspecific hybridization involving stochastic protoplast fusion within these shuffled strains. By combining genome shuffling and interspecific hybridization, we constructed a hybrid designated FEEL-1 with more than 3-fold improvements in ε-PL production and demonstrated a new strategy to improve phenotype of industrial strains.
Materials and Methods
Materials
Lysozyme, phosphoenolpyruvate (PEP), malate dehydrogenase (MD), ATP, ADP, NADH, NADP, DTNB, 6-phosphate dehydrogenase, acetyl CoA, L-lactate dehydrogenase, and oxalacetic acid were purchased from Sigma (USA); polyethylene glycol (PEG 6000) were purchased from Sinopharm Chemical Reagent Co. Ltd (China); arbitrary primers for random amplified polymorphic DNA (RAPD) were purchased from Sangon Biotech (Shanghai) Co., Ltd.; all other chemicals used in this work were of analytical grade.
Organism and Growth Media
S. padanus, S. griseofuscus, S. graminearus, S. hygroscopicus, and S. albulus were isolated from soil samples collected in Wuxi City, China. Shuffled strains of these species were obtained by three rounds of genome shuffling. FEEL-1 was obtained by stochastic interspecific hybridization among the shuffled strains. These colonies were grown on BTN solid medium consisted of glucose (10 g), yeast extract (2 g), peptone (4 g), agar (20 g) per liter deionized water. pH was adjusted to 7.2 before sterilization. Sporulation emerged on BTN agar plates after incubation for 7 days at 30 °C.
Screening plates were BTN agar supplemented with different concentrations of GLU or SG and SA agar contained sodium succinic acid 50 g, (NH4)2SO4 15 g, KH2PO4 1 g, K2HPO4 3 g, and agar 20 g per liter deionized water.
Regeneration medium (RM) was BTN supplemented with sucrose (103 g/L), MgCl2·6H2O (10 g/L), and CaC12·2H2O (0.4 g/L). pH was adjusted to 7.2 with NaOH (1 N) after sterilization. Protoplasts would grow on RM agar plates and generate spores after incubation for 10 days at 30 °C.
Liquid medium contained soluble starch (10 g), yeast extract (6 g), MgSO4·7H2O (1.0 g), K2HPO4 (2.0 g), and KH2PO4 (0.5 g) per liter deionized water. pH was adjusted to 7.2 with NaOH (1 N) before sterilization. Spores of strains were inoculated into this medium and incubated at 30 °C on a rotating shaker. After 24 h of incubation, mycelia were obtained to prepare protoplasts.
Fermentation medium (M3G) consisted of glucose (50 g), (NH4)2SO4 (10 g), yeast extract (5 g), MgSO4·7H2O (0.5 g), K2HPO4 (0.8 g), KH2PO4 (1.4 g), ZnSO4·7H2O (0.04 g), and FeSO4·7H2O (0.03 g) per liter deionized water. pH was adjusted to 6.8 with NaOH (1 N) before sterilization.
Strain Mutagenesis and Mutant Screening
Wild-type strains mutagenized with NTG to obtain the initial mutant library was as follows: spores (1 × 108–5 × 108) were suspended in 10 ml of sterilized 0.9 % saline water and then harvested by centrifugation (4,000×g, Sigma, USA). The harvested spores were washed twice with 5 ml of sterilized 10 mM Tris–HCl (pH 7.5) and then treated with 5 ml of 0.01 % NTG in 10 mM Tris–HCl buffer (pH 7.5) buffer for 40 min to give a survival rate of 1.0 %. The spores were then spread onto the screening plates respectively. UV irradiation was performed by exposing colonies on screening plates directly to an UV light (power of 8 W) at a distance of 20 cm for 3 min, with the same survival rate as NTG mutation. These plates were incubated at 30 °C in incubator. The fast grown colonies were picked out for ε-PL production analysis in shake flask. The mutants that showed higher levels of ε-PL production over wild type were selected as the starters for genome shuffling.
Genome Shuffling
Genome shuffling was carried out using modified described methods [7, 10, 12]. Starters isolated from NTG-treated and UV-treated strains were respectively inoculated into liquid medium at 30 °C for 24 h. Cells were harvested by centrifugation at 4,000×g for 5 min at 4 °C, washed twice with 30 ml of PB buffer [20 mM Tris–HCl buffer (pH 7.5) supplemented with 103 g/L sucrose and 2 g/L MgCl2], and treated with lysozyme (5 mg/ml in PB buffer) at 30 °C for 90 min. After observation of protoplast formation under a compound light microscope (Sigma, USA), equal number of protoplasts from different populations were mixed and divided equally into two parts. One part was inactivated with UV irradiation for 60 min (power of 8 W), and the other was heat treated at 70 °C for 30 min [13]. The survival frequency of protoplasts was zero in these two inactivation methods. Both inactivated protoplasts were mixed in a cell ratio of 1:1, centrifuged, and resuspended in 50 μl PB buffer. Protoplasts were fused by suspension in 5 ml of PB buffer containing 40 % PEG 6000, and 10 mM CaCl2. After gentle shaking for 10 min at 38 °C, the suspension was diluted 5-fold with PB buffer and spread on RM agar plates. Recombinants grew on RM agar plates and generated spores after incubation for 10 days at 30 °C. Subsequent rounds of genome shuffling were carried out by repeating the protoplast fusion described above.
Interspecific Hybridization by Protoplast Fusion
The purified spores of all shuffled strains were inoculated into liquid medium respectively and incubated at 30 °C on a rotating shaker. After 24 h of incubation, mycelia were obtained to prepare protoplasts. Interspecific hybridization included two-species cross and five-species cross by mixing different species of protoplasts together. Process of protoplast fusion was the same as genome shuffling.
Shake Flask and Bioreactor Cultivation
Wild-type strains, shuffled strains, and hybrids were transferred respectively into shake flasks containing M3G medium in orbital shakers at 30 °C, 200 rpm, and maintained up to 96 h. Each strain was grown in three separate shake flasks, and the mean values were reported.
A 5-l fermentor (Shanghai Baoxing Bio-engineering Equipment Co., Ltd., China) containing 2,760 ml of M3G was inoculated with 240 ml of 24-h-old seed culture. The initial pH was adjusted to 6.8 with NaOH (2 N) and change of pH during cultivation was detected by a pH electrode (Mettler Toledo Company, Switzerland). When pH naturally dropped to 3.8 (suitable pH for ε-PL production), NH4OH solution was automatically added to the culture broth to maintain it. Fed-batch fermentation was carried out by adding mixture of glucose and ammonium sulfate whose proportion was the same as the initial liquid medium. When glucose concentration decreased to 0.5 %, the mixture was manually added to the broth until the final glucose concentration reached 1.5 %.
The culture broth was sampled every 6 h, centrifuged, and the resulting supernatant was used for measurement of ε-PL and residual glucose concentrations. The concentration of ε-PL in various solutions was measured according to the method of Itzhaki’s [14] and Shima’s [15] with modifications; 0.5 ml of sample solution was mixed with 2 ml methyl orange (MeO) solution (1 mM MeO and 50 mM sodium phosphate, pH 7.0) and vortexed. After incubation for 30 min at 30 °C, mixtures were centrifuged at 4,000×g, and the resulting supernatants were diluted 10-fold. Absorbance of the resulting dilutions was measured at 465 nm. The biomass was determined by harvesting culture sample, filtering, washing the mycelia twice with distilled water, and drying until a constant weight was achieved at 100 °C. The glucose concentration was determined by a biosensor analyzer SBA-40D (Shandong Academy of Sciences).
Identification of Hybrids Using RAPD
Genomic DNA from Streptomyces was isolated using a test kit for extracting bacterial genome (Sangon Biotech Co., Ltd.).Twenty-six arbitrary primers were used in RAPD to distinguish FEEL-1 from five species of parents, and three of them were proved available: primer S7 (ACAACGCCTC); primer S12 (AGGGCCGTCT); primer S15 (GGGTACCCGT). Amplification products were analyzed by electrophoresis in 2.0 % agarose gels in 1× TBE buffer and detected after staining with GoldView [16].
Comparison for Enzyme Activities Between FEEL-1 and Its Parents
Spores of FEEL-1 and its three parents were inoculated into M3G medium and incubated at 30 °C, 200 rpm on a rotating shaker, respectively. After 30 h of incubation, 150 ml mycelium of each strain was obtained to prepare cell extracts. Enzyme activities consisted of hexokinase (HK), phosphoenolpyruvate carboxylase (PEPC), aspartokinase (AK), and citrate synthase (CS) were measured according to Teichgraber, Murmu, Hamano, and James, respectively [17–20].
Results
Genome Shuffling Improved ε-PL Production
The productivity of ε-PL in wild-type strains of S. padanus, S. griseofuscus, S. graminearus, S. hygroscopicus, and S. albulus was low, about 0.38–0.49 g/L in shake flask. In our previous work, genome shuffling had been employed to improve ε-PL production as 0.8 g/L by glucose tolerance in S. graminearus [9]. In this research, we adopted genome shuffling to increase ε-PL productivity of these five wild-type strains by GLU tolerance, SG tolerance, and SA as sole carbon source, which were likely to conduce to higher enzyme activities of HK, AK, and PEPC, respectively [17, 19, 21].
We took S. padanus by SG tolerance as a model to illustrate the process of genome shuffling. For SG-tolerance of wild type (WT), the concentration of 20 g/L was its threshold and strains could hardly grow on BTN agar containing corresponding dose of SG. Consequently, we generated an initial library of S. padanus mutants from BTN agar containing 22 g/L SG and selected those mutants that showed higher levels of ε-PL production over WT. NTG and UV irradiation mutagenesis were introduced to generate the first population of SG-tolerant variants of WT. Ninety-six colonies showing rapid growth were picked out for shake flask fermentation test and six strains exhibited higher level of ε-PL production (0.60 ± 0.01 g/L) over WT (0.46 g/L). The protoplasts of these six mutants were subjected to the first round of genome shuffling. Forty-six colonies showing rapid growth on plates with higher concentration of SG (24 g/L) were picked out for shake flask fermentation test and six colonies were identified as higher ε-PL producers (0.68 ± 0.01 g/L) compared with mutated parents. These six strains were then adopted for the second round of genome shuffling and six colonies out of forty-two were selected from the shuffled library which could grow on BTN plates containing 26 g/L of SG. These six strains produced 0.75 ± 0.01 g/L of ε-PL, which were subjected to the third round of genome shuffling. The resulting third round population of 40 isolates grown fast on BTN plates containing SG 28 g/L was screened for ε-PL productivity. Six isolates were found to exhibit ε-PL productivities of 0.81 ± 0.01 g/L and the whole process of genome shuffling was shown in Fig. 1.
Process of genome shuffling of S. padanus by SG-tolerance. WT wild type of S. padanus, UV/NTG mutant strains, F1 strains from the first round of genome shuffling, F2 strains from the second round of genome shuffling, F3 strains from the third round of genome shuffling. Dotted line represents the ε-PL production level of S. padanus
By following the above example, we obtained 41 shuffled strains in total including three kinds of tolerance among five species of strains (Table 1), with ε-PL production ranging from 0.75 to 0.81 g/L compared with 0.38–0.49 g/L of wild types in shake flask. However, extensive improvement in ε-PL production failed after three rounds of genome shuffling in our study. We hypothesized the main reason was that the improvement of one tolerance could give rise to higher activity of one specific enzyme (HK or PEPC or AK), and the resulting incoordination for integral metabolism in shuffled strains limited a further increase of ε-PL production. Therefore, we considered making a combination of these shuffled strains through stochastic protoplast fusion. In fact, this hypothesis was inspired by Effendi Leonard, who achieved an overproduction of terpenoid in Escherichia coli combining metabolic and protein engineering [22].
Interspecific Hybridizations Between Two Species
First we reported the results of attempted protoplast fusions between two species of all possible hybridizations among shuffled strains (for example, 11 shuffled strains of S. padanus fused with eight of S. griseofuscus, Table 1), involving ten kinds of pairing combinations. Surprisingly, all combinations were proved to produce fusion by inactivating one or two species. For example, S. griseofuscus could secrete melanin while S. hygroscopicus could not (Fig. 2a), but inactivated protoplasts of S. griseofuscus treated by heat were able to fuse with live ones of S. hygroscopicus, forming hybrids which could secrete melanin again (Fig. 2b); inactivated protoplasts of S. padanus and S. albulus treated by UV and heat lost their regeneration ability, but it could contribute to formation of live hybrids when fused them together, exhibiting different morphologies. From Fig. 2c, we could see that colonies of S. padanus on RM plates were orbicular and yellow, while S. albulus showed irregular shape and yellow-white, but a third colonies appeared after interspecific hybridization with bigger size, orbicular, and milky white. We considered these strains with different morphologies as hybrids. However, it is a pity that no significant improvements for ε-PL production were observed in these two-species hybridizations (data not shown).
Morphologies of some parent strains and hybrids in two-species hybridizations. a S. hygroscopicus (1) could not secret pigment and S. griseofuscus (2) could secret melanin; b dead (inactivated by heat) protoplasts of S. griseofuscus were able to fuse with live ones of S. hygroscopicus, forming hybrids which could secrete melanin again; c protoplast fusion between S. albulus (1) and S. padanus (2) produced hybrids (3) with different morphologies
Interspecific Hybridization Among Five Species
Two-species hybridizations failed to give hybrids with improved ε-PL production probably because the lower number of shuffled strains in two species. Thus we carried out an interspecific hybridization involving 41 shuffled strains among five species of the Streptomyces group. This operation was (1) to assess the possibility of obtaining hybrids with significant improvement for ε-PL production, whose primary purpose was the same as two-species hybridizations and (2) to investigate the possibility of hybridization involving multiple species because enough quantity of strains could no doubt increase the probability of interspecific hybridization.
Process of protoplast fusion was photographed by a photomicrography instrument shown in Fig. 3. Under induction of PEG, protoplasts aggregated and fused together owing to fluidity of the cell membrane. Two-cell fusion, three-cell, four-cell, and multiple-cell could be clearly seen, but which species took part in a fusion was visually unknown. Protoplasts fusion is a complicated process of chromosome hybridization, some could be fused from the whole genome, forming diploids or polyploids [21, 23]; some could be fused from certain genes [21, 24] and some could not be fused [21, 25], forming only heterokaryons. After several generations, heterokaryons differentiate into their parents separately and some hybrids may gradually discard certain useless genes, turning into stable recombinants.
Owing to absence of adequate genetic markers, we preliminary distinguished hybrids from parent species by morphology and spores color on RM plates. These colonies were then streaked on BTN media to sporulate, displaying different colors. For example, S. griseofuscus was gray; S. hygroscopicus was brown; S. albulus was brown-black; two of the hybrids were gray-black and dark-green (Fig. 4). Some other colonies with similar morphology or spores colors were not shown.
In hybridization among five species, 48 hybrids showing rapid growth were picked out for shake flask fermentation test and one strain designated FEEL-1 was identified as a higher ε-PL producer (1.12 g/L) compared with the shuffled strains (0.75–0.81 g/L; Fig. 5). Subsequently, it was proved that FEEL-1 was stable after eight generations of subcultures, with ε-PL production between 1.07 and 1.15 g/L in shake flask. This result implied that the hybrid FEEL-1 had good reusability and genomic stability.
ε-PL production of FEEL-1 in shake flask was higher than those in wild-type strains and shuffled strains. WT five species of wild-type strains, S1–S41 41 shuffled strains obtained by genome shuffling, FEEL-1 a hybrid with the highest ε-PL production. Dotted lines represent the obvious gradients for ε-PL production
RAPD
Although FEEL-1 was a hybrid obtained by protoplast fusion from 41 shuffled strains, including five different species, it was a slight probability incident that all species were involved in the defacto hybridization. Thus, RAPD was adopted to confirm the parent species of FEEL-1. Twenty-six arbitrary primers were employed in this research and three of them showed specificity. RAPD fingerprints of FEEL-1 and the five possible parent species using different primers were shown in Fig. 6. For each primer evaluated, one to six DNA fragments were amplified and the sizes of fragments ranged from 150 to 750 bp. All the major fragments amplified by RAPD were reproducible under identical conditions. Using primer S7, one fragment of 250 bp and another of 150 bp were present in S. albulus and S. padanus, respectively. And these two specific fragments appeared in FEEL-1 simultaneously (Fig. 6a), indicating that S. albulus and S. padanus were two possible parents of FEEL-1. Likewise, when primer S12 was used, the result showed that S. albulus and S. griseofuscus were involved in hybridization (Fig. 6b) while primer S15 only displayed S. padanus was one of parents for FEEL-1 (Fig. 6c). In summary, RAPD results indicated that possibly three species (S. griseofuscus, S. padanus, and S. albulus) participated in the defacto hybridization. Here we must notice a fact that these three species were consisted of 29 shuffled strains including different tolerances (Table 1), and we were only able to make clear which species were introduced into FEEL-1 but not certain specific shuffled strains.
RAPD results preliminary indicated that three species (S. griseofuscus; S. padanus; S. albulus) participated in the defacto hybridization. a–c RAPD fingerprints of primer S7, S12, and S15, respectively; 0–6 marker, S. graminearus, S. hygroscopicus, S. albulus, S. griseofuscus, S. padanus, and FEEL-1, respectively
Fed-Batch Fermentation
Although ε-PL production of FEEL-1 was higher than those in wild-type strains and shuffled strains in shake flask (Fig. 5), our expectation was that FEEL-1 could take advantage of its high productivity in large-scale fermentation to meet industrial needs. Therefore behaviors of FEEL-1 and its three parent species (including shuffled strains and wild-type strains) were detected for comparison in a 5-l fermentation tank. Considering large amount of parent strains (29 shuffled strains) and reduction of workload, we selected three of them for scale-up fermentation: S. griseofuscus (GLU tolerance), S. padanus (SA as sole carbon source), and S. albulus (SG tolerance). The fermentation results of these strains were shown in Fig. 7. For S. griseofuscus, S. padanus, and S. albulus, ε-PL productions of the wild-type strains were 7.2, 7.5, and 8.1 g/L, respectively. But the shuffled strains of corresponding species exhibited higher ε-PL yields as 15.5, 13.5, and 16.9 g/L, respectively. Therefore, the differences of ε-PL production between shuffled strains and wild-type strains were obvious. Besides, consistent with the results of five-species hybridization in shake flask fermentation, the hybrid FEEL-1 produced much more ε-PL as 24.5 g/L, about 63–81 % higher over those of shuffled ones. More importantly, because biomass of these strains expressed no obvious difference (23–26 g/L), it was safe to say that the improvement of ε-PL in FEEL-1 was independent on the biomass but due to the stronger ε-PL synthetic ability, which proved the effectiveness of genome shuffling along with interspecific hybridization as a new breeding strategy.
Comparison for ε-PL and biomass production between FEEL-1 and its three parents in fed-batch fermentation. Mixture of glucose and ammonium sulfate was fed into fermentor under pH 3.8. When glucose concentration decreased to 0.5 %, the mixture was manually added to the broth until the final glucose concentration reached 1.5 %. The whole fermentation period was 168 h
Comparison for Enzyme Activities Between FEEL-1 and Its Parents
In order to give a preliminary explanation why shuffled strains and FEEL-1 exhibited more ε-PL productivity, enzyme activities involving HK, PEPC, and AK were investigated (Table 2). Take S. griseofuscus for example, several enzyme activities in WT were low but HK activity in shuffled strain by GLU tolerance showed 6-fold improvements, indicating a more active glycolytic pathway. This result was the same as S. graminearus in our previous work [9]. Similarly, for S. padanus (SA) and S. albulus (SG), the shuffled strains respectively exhibited higher enzyme activities in PECP and AK. Although improvements of enzyme activities proved the efficiency of genome shuffling, the resulting incoordination for integral metabolism in shuffled strains limited a further increase of ε-PL production. But after the interspecific hybridization, situation was found to be better. As shown in Table 2, despite HK activity in FEEL-1 was far lower than that in S. griseofuscus (GLU) it was higher than those in other two parents. This result suggested that glycolytic pathway (EMP) may be not the limiting step for ε-PL production. While enzyme activities for PEPC and AK in FEEL-1 were obviously higher than those in shuffled parents, it indicated a success of interspecific hybridization and meanwhile suggested that diaminopimelic acid (DPA) pathway limit the ε-PL production. In bacteria, lysine is synthesized through DPA pathway. DPA is formed via aspartic acid produced by combining oxaloacetate (OXA) with the ammonium ion of a nitrogen source, where AK plays an important role in this process. Meanwhile, two following metabolic pathways might increase the source of OXA. The carbon dioxide fixation ability in strain FEEL-1 was enhanced because PEPC catalyzed a reaction from PEP to OXA [26]. On the other hand, higher CS activity (Table 2) in FEEL-1 strengthened the TCA circle, leading greater flux to DPA pathway. This result was some what similar to an ε-PL producing strain USE-51, who was reported that adding citrate to medium could facilitate the ε-PL production [27].
Discussion
Efforts to increase target products in industrial strains previously focused on (1) overexpression of pathway enzymes by metabolic engineering, and (2) optimizing the expression of enzymes by codon bias. However, these approaches are able to be realized only on premise that the details of biosynthetic pathway and regulatory mechanism of organisms are characterized. Unfortunately, for our wild-type strains screened from soils, it is impractical to obtain mutants with remarkably improved ε-PL production in short period by these rational methods owing to lack of genetic backgrounds. In this research, we employed genome shuffling by different tolerances to improve enzyme activities of HK, PEPC, or AK respectively thus increased the ε-PL production. But it was still limited by other inherent low-enzyme activities and we subsequently ameliorated this incoordination combining with interspecific hybridization. Recombination within a selected population amplifies the genetic diversity of the population by creating new mutant combinations, and can thereby improve the performance of individuals within the population [12]. Besides, cellular phenotypes are by nature complex, and result from the dynamic interaction of genes and gene products distributed throughout a cell's genome. Therefore, it is difficult to clearly elucidate which genes were responsible for the enhancement of enzyme activities. What is more, which useful genes from the parent strains were introduced into FEEL-1 needs further investigation because most primers employed in this research showed no specificity.
Our approach highlights the importance of genome shuffling along with interspecific hybridization as a new breeding strategy for improving phenotype of industrial strains. By this strategy, a hybrid FEEL-1 with ε-PL production of 24.5 g/L was obtained, which was more than 3-fold higher compared with wild-type strains. Although a good result was achieved, there are also certain disparities compared to the reported high ε-PL production of 34–48 g/L [4–6]. We suspect that doing some fermentation studies might give significant yield improvement, such as optimization of culture medium and fermentation process. These works are underway in our laboratory.
References
Hiraki, J., Ichikawa, T., & Ninomiya, S. (2003). Use of ADME studies to confirm the safety of polylysine as a preservative in food. Regul Toxicol Pharm, 37, 328–340.
Shih, I. L., & Shen, M. H. (2006). Microbial synthesis of poly (ε-lysine) and its various applications. Bioresource Technology, 97, 1148–1159.
Li, S., Tang, L., & Chen, X. S. (2010). Isolation and characterization of a novel ε-poly-l-lysine producing strain: streptomyces griseofuscus. Journal of Industrial Microbiology and Biotechnology, 38, 563–577.
Kahar, P., & Iwata, T. (2001). Enhancement of ε-polylysine production by Streptomyces albulus strain 410 using pH control. Journal of Bioscience and Bioengineering, 91, 190–194.
Zhang, Y., Feng, X. H., & Xu, H. (2010). ε-poly-l-lysine production by immobilized cells of Kitasatospora sp. MY 5–36 in repeated fed-batch cultures. Bioresource Technology, 101, 5523–5527.
Chen, X. S., Ren, X. D., & Dong, N. (2012). Culture medium containing glucose and glycerol as a mixed carbon source improves e-poly-l-lysine production by Streptomyces sp. M-Z18. Bioprocess and Biosystems Engineering, 35, 469–475.
Dai, M. H., & Copley, S. D. (2004). Genome shuffling improves degradation of the anthropogenic pesticide pentachlorophenol by Sphingobium chlorophenolicum ATCC 39723. Applied and Environmental Microbiology, 70, 2391–2397.
Hida, H., Yamada, T., & Yamada, Y. (2007). Genome shuffling of Streptomyces sp. U121 for improved production of hydroxycitric acid. Applied Microbiology and Biotechnology, 73, 1387–1393.
Li, S., Li, F., & Chen, X. S. (2012). Genome shuffling enhanced ε-Poly-l-Lysine production by improving glucose tolerance of Streptomyces graminearus. Applied Biochemistry and Biotechnology, 166, 414–423.
Patnaik, R., Louie, S., Gavrilovic, V., Perry, K., Stemmer, W. P. C., Ryan, C. M., et al. (2002). Genome shuffling of Lactobacillus for improved acid tolerance. Nature Biotechnology, 20, 707–712.
Yu, L., Pei, X. L., & Lei, T. (2008). Genome shuffling enhanced L-lactic acid production by improving glucose tolerance of Lactobacillus rhamnosus. Journal of Biotechnology, 134, 154–159.
Zhang, Y. X., Perry, K., & Vincy, A. (2002). Genome shuffling leads to rapid phenotypic improvement in bacteria. Natural Letters, 415, 644–646.
Hopwood, D. A., & Wright, H. M. (1979). Factors affecting recombinant frequency in protoplast fusion of Streptomyces coelicolor. Journal of General Microbiology, 111, 137–143.
Itzhaki, R. F. (1972). Colorimetric method for estimating poly-lysine and polyarginine. Analytical Biochemistry, 50, 569–574.
Shima, S., & Sakai, H. (1981). Poly-l-lysine produced by Streptomyces. Part II. taxonomy and fermentation studies. Agricultural and Biological Chemistry, 45, 2503–2508.
Welsh, J., & McClelland, M. (1990). Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Research, 18, 7213–7218.
Hamano, Y., & Nicchu, I. (2007). ε-Poly-l-lysine producer, Streptomyces albulus, has feedback-inhibition resistant aspartokinase. Applied Microbiology and Biotechnology, 76, 873–882.
James, C. K., & Liang, B. B. (1994). Brain mitochondrial citrate synthase and glutamate dehydrogenase: differential inhibition by fatty acyl coenzyme a derivatives. Metabolic Brain Disease, 9, 143–152.
Murmu, J., & William, C. (2007). Phosphoenolpyruvate carboxylase protein kinase from developing castor oil seeds: partial purification, characterization, and reversible control by photosynthate supply. Planta, 226, 1299–1310.
Teichgraber, P., & Biesold, D. (1972). Subcellular localization of hexokinase in the rat cortex. Biokhimiya, 37, 748–756.
Ferenc, K., & Peberdy, J. (1984). Further studies on protoplast fusion and interspecific hybridization within the Aspergillus nidulans group. Journal of General Microbiology, 130, 2229–2236.
Leonard, E., Kumaran, P., & Thayer, K. (2010). Combining metabolic and protein engineering of a terpenoid biosynthetic pathway for overproduction and selectivity control. Proc Natl Acad Sci, 107, 13654–13659.
Hohzoh, K., & Toshiro, W. (1990). Intergeneric hybridization between Monascus anka and Asperyillus oryzae by protoplast fusion. Applied Microbiology and Biotechnology, 33, 671–676.
Nalin, R., & David, H. (1983). Spheroplast fusion as a mode of genetic recombination in mycobacteria. Journal of General Microbiology, 129(1227–1), 237.
John, F., & Hendrik, E. (1977). Interspeeifie hybridization between Penicillium chlysogenum and Penicillium cyaneofulvum following protoplast fusion. Molec gen Genet, 157, 281–284.
Helena, B., & Hugh, G. (1993). Phosphoenolpyruvate carboxylase from Streptomyces coelicolor A3(2): purification of the enzyme, cloning of the ppc gene and over-expression of the protein in a streptomycete. The Biochemical Journal, 293, 131–136.
Hirohara, H., Takehara, M., & Saimura, M. (2006). Biosynthesis of poly(ε-l-lysine)s in two newly isolated strains of Streptomyces sp. Applied Microbiology and Biotechnology, 73, 321–331.
Acknowledgments
This work was supported by a grant from Wuxi Science and Technology Support Program (CYE21N1107), the Priority Academic Program Development of Jiangsu Higher Education Institutions, and 111 Project (111-2-06).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Li, S., Chen, X., Dong, C. et al. Combining Genome Shuffling and Interspecific Hybridization Among Streptomyces Improved ε-Poly-l-Lysine Production. Appl Biochem Biotechnol 169, 338–350 (2013). https://doi.org/10.1007/s12010-012-9969-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-012-9969-0