Abstract
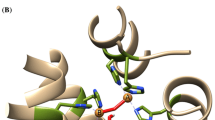
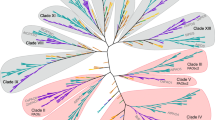
Polyphenol oxidases (PPOs) are widely distributed enzymes among animals, plants, bacteria, and fungi. PPOs often have significant role in many biologically essential functions including pigmentation, sclerotization, primary immune response, and host defense mechanisms. In the present study, forty-seven full-length amino acid sequences of PPO from bacteria, fungi, and plants were collected and subjected to multiple sequence alignment (MSA), domain identification, and phylogenetic tree construction. MSA revealed that six histidine, two phenylalanine, two arginine, and two aspartic acid residues were highly conserved in all the analyzed species, while a single cysteine residue was conserved in all the plant and fungal PPOs. Two major sequence clusters were constructed by phylogenetic analysis. One cluster was of the plant origin, whereas the other one was of the fungal and bacterial origin. Motif GGGMMGDVPTANDPIFWLHHCNVDRLWAVWQ was found in all the species of bacterial and fungus sources. In addition, seven new motifs which were unique for their group were also identified.




Similar content being viewed by others
References
Mayer, A. M. (2006). Phytochemistry, 67, 2318–2331.
Mason, H. (1955). Adv Enzymol, 16, 105–184.
Umit, U. M. (2007). Food Chem, 100, 909–913.
Wichers, H. J., Recourt, K., Hendriks, M., Ebbelaar, C. F. M., Biancone, G., & Hoeberichts, F. A. (2003). Appl Microbiol Biotechnol, 61, 336–341.
Bailey, T. L., & Elkan, C. (1995). Mach Learn, 21(1–2), 51–80.
Bailey, T. L., & Gribskov, M. (1998). Bioinformatics, 14(1), 48–54.
Tamura, K., Dudley, J., Nei, M., & Kumar, S. (2007). Mol Biol Evol, 24, 1596–1599.
Marusek, C. M., Trobaugh, N. M., Flurkey, W. H., & Inlow, J. K. (2006). J Inorg Biochem, 100, 108–123.
Garcia-Borron, J. C., & Solano, F. (2002). Pigment Cell Res, 15, 162–173.
van Gelder, C. W. G., Flurkey, W. H., & Wichers, H. J. (1997). Phytochemistry, 45, 1309–1323.
Gerdemann, C., Eicken, C., & Krebs, B. (2002). Acc Chem Res, 35, 183–191.
Klabunde, T., Eicken, C., Sacchettini, J. C., & Krebs, B. (1998). Nat Struct Biol, 5, 1084–1090.
Eickena, C., Krebs, B., & Sacchettini, J. C. (1999). Curr Opin Struct Biol, 9, 677–683.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Malviya, N., Srivastava, M., Diwakar, S.K. et al. Insights to Sequence Information of Polyphenol Oxidase Enzyme from Different Source Organisms. Appl Biochem Biotechnol 165, 397–405 (2011). https://doi.org/10.1007/s12010-011-9259-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-011-9259-2