Abstract
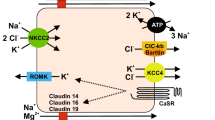
Calcium nephrolithiasis is a common condition. Family-based genetic linkage studies and genome-wide association studies (GWASs) have uncovered a run of important candidate genes involved in renal Ca++ disorders and kidney stone diseases. The susceptible genes include NKCC2, ROMK and ClCkb/Barttin that underlie renal salt excretion; claudin-14, -16 and -19 that underlie renal Ca++ excretion; and CaSR that provides a sensing mechanism for the kidney to regulate salt, water and Ca++ homeostasis. Biological and physiological analyses have revealed the cellular mechanism for transepithelial Ca++ transport in the kidney that depends on the concerted action of these gene products. Although the individual pathogenic weight of the susceptible genes in nephrolithiasis remains unclear, perturbation of their expression or function compromises the different steps within the integrated pathway for Ca++ reabsorption, providing a physiological basis for diagnosing and managing kidney stone diseases.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Coe FL, Evan A, Worcester E. Kidney stone disease. J Clin Invest. 2005;115:2598–608.
Hess B, Hasler-Strub U, Ackermann D, et al. Metabolic evaluation of patients with recurrent idiopathic calcium nephrolithiasis. Nephrol Dial Transplant. 1997;12:1362–8.
Thorleifsson G, Holm H, Edvardsson V, et al. Sequence variants in the CLDN14 gene associate with kidney stones and bone mineral density. Nat Genet. 2009;41:926–30.
Friedman PA. Renal calcium metabolism. In Seldin and Giebisch’s The Kidney – Physiology and Pathophysiology. 2008; volume 2; chapter 65; p1851-1890.
Mensenkamp AR, Hoenderop JG, Bindels RJ. Recent advances in renal tubular calcium reabsorption. Curr Opin Nephrol Hypertens. 2006;15:524–9.
Liebman SE, Taylor JG, Bushinsky DA. Idiopathic hypercalciuria. Curr Rheumatol Rep. 2006;8:70–5.
Lamberg BA, Kuhlback B. Effect of chlorothiazide and hydrochlorothiazide on the excretion of calcium in urine. Scand J Clin Lab Invest. 1959;11:351–7.
Nijenhuis T, Vallon V, van der Kemp AW, et al. Enhanced passive Ca2+ reabsorption and reduced Mg2+ channel abundance explains thiazide-induced hypocalciuria and hypomagnesemia. J Clin Invest. 2005;115:1651–8.
Muto S, Hata M, Taniguchi J, et al. Claudin-2-deficient mice are defective in the leaky and cation-selective paracellular permeability properties of renal proximal tubules. Proc Natl Acad Sci U S A. 2010;107:8011–6. This article reports the renal phenotypes of claudin-2 knockout mice and emphasizes the role of claudin-2 channel in paracellular Ca ++ reabsorption in the proximal tubule of the kidney.
Hou J, Goodenough DA. Claudin-16 and claudin-19 function in the thick ascending limb. Curr Opin Nephrol Hypertens. 2010;19:483–8. This article is a recent review of claudin physiology in the thick ascending limb of the kidney.
Hou J, Renigunta A, Gomes AS, et al. Claudin-16 and claudin-19 interaction is required for their assembly into tight junctions and for renal reabsorption of magnesium. Proc Natl Acad Sci U S A. 2009;106:15350–5.
Hou J, Renigunta A, Konrad M, et al. Claudin-16 and claudin-19 interact and form a cation-selective tight junction complex. J Clin Invest. 2008;118:619–28.
Hou J, Shan Q, Wang T, et al. Transgenic RNAi depletion of claudin-16 and the renal handling of magnesium. J Biol Chem. 2007;282:17114–22.
Hou J, Paul DL, Goodenough DA. Paracellin-1 and the modulation of ion selectivity of tight junctions. J Cell Sci. 2005;118:5109–18.
Farquhar MG, Palade GE. Junctional complexes in various epithelia. J Cell Biol. 1963;17:375–412.
Miller F. Hemoglobin absorption by the cell of the proximal convoluted tubule in mouse kidney. J Biophys Biochem Cytol. 1960;8:689–718.
Furuse M, Hirase T, Itoh M, et al. Occludin - a novel integral membrane protein localizing at tight junctions. J Cell Biol. 1993;123:1777–88.
Ebnet K, Suzuki A, Ohno S, et al. Junctional adhesion molecules (JAMs): more molecules with dual functions? J Cell Sci. 2004;117:19–29.
Lal-Nag M, Morin PJ. The claudins. Genome Biol. 2009;10:235.1–7.
Mineta K, Yamamoto Y, Yamazaki Y, et al. Predicted expansion of the claudin multigene family. FEBS Lett. 2011;585:606–12.
Krause G, Winkler L, Mueller SL. Structure and function of claudins. Biochim Biophys Acta. 2008;1778:631–45.
Colegio OR, Van Itallie CM, Rahner C, et al. Claudin extracellular domains determine paracellular charge selectivity and resistance but not tight junction fibril architecture. Am J Physiol Cell Physiol. 2003;284:C1346–54.
Alexandre MD, Jeansonne BG, Renegar RH, et al. The first extracellular domain of claudin-7 affects paracellular Cl− permeability. Biochem Biophys Res Commun. 2007;357:87–91.
Van Itallie CM, Anderson JM. Claudins and epithelial paracellular transport. Annu Rev Physiol. 2006;68:403–29.
Cukierman L, Meertens L, Bertaux C, et al. Residues in a highly conserved claudin-1 motif are required for hepatitis C virus entry and mediate the formation of cell-cell contacts. J Virol. 2009;83:5477–84.
Fujita K, Katahira J, Horiguchi Y, et al. Clostridium perfringens enterotoxin binds to the second extracellular loop of claudin-3, a tight junction integral membrane protein. FEBS Lett. 2000;476:258–61.
Hamazaki Y, Itoh M, Sasaki H, et al. Multi-PDZ-containing protein 1 (MUPP1) is concentrated at tight junctions through its possible interaction with claudin-1 and junctional adhesion molecule (JAM). J Biol Chem. 2001;277:455–61.
Itoh M, Furuse M, Morita K, et al. Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J Cell Biol. 1999;147:1351–63.
Van Itallie C, Rahner C, Anderson JM. Regulated expression of claudin-4 decreases paracellular conductance through a selective decrease in sodium permeability. J Clin Invest. 2001;107:1319–27.
Colegio OR, Van Itallie CM, McCrea HJ, et al. Claudins create charge-selective channels in the paracellular pathway between epithelial cells. Am J Physiol Cell Physiol. 2002;283:C142–7.
Ben-Yosef T, Belyantseva IA, Saunders TL, et al. Claudin 14 knockout mice, a model for autosomal recessive deafness DFNB29, are deaf due to cochlear hair cell degeneration. Hum Mol Genet. 2003;12:2049–61.
Yu AS, Enck AH, Lencer WI, et al. Claudin-8 expression in MDCK cells augments the paracellular barrier to cation permeation. J Biol Chem. 2003;278:17350–9.
Wen H, Watry DD, Marcondes MC, et al. Selective decrease in paracellular conductance of tight junctions: role of the first extracellular domain of claudin-5. Mol Cell Biol. 2004;24:8408–17.
Furuse M, Furuse K, Sasaki H, et al. Conversion of zonulae occludentes from tight to leaky strand type by introducing claudin-2 into Madin-Darby canine kidney I cells. J Cell Biol. 2001;153:263–72.
Van Itallie CM, Fanning AS, Anderson JM. Reversal of charge selectivity in cation or anion selective epithelial lines by expression of different claudins. Am J Physiol Renal Physiol. 2003;285:F1078–84.
Tang VW, Goodenough DA. Paracellular ion channel at the tight junction. Biophys J. 2003;84:1660–73.
Tsukita S, Furuse M. Pores in the wall. Claudins constitute tight junction strands containing aqueous pores. J Cell Biol. 2000;149:13–6.
Watson CJ, Rowland M, Warhurst G. Functional modeling of tight junctions in intestinal cell monolayers using polyethylene glycol oligomers. Am J Physiol Cell Physiol. 2001;281:C388–97.
Van Itallie CM, Holmes J, Bridges A, et al. The density of small tight junction pores varies among cell types and is increased by expression of claudin-2. J Cell Sci. 2008;121:298–305.
Yu AS, Cheng MH, Angelow S, et al. Molecular basis for cation selectivity in claudin-2-based paracellular pores: identification of an electrostatic interaction site. J Gen Physiol. 2009;133(1):111–27.
Simon DB, Karet FE, Hamdan, et al. Bartter’s syndrome, hypokalaemic alkalosis with hypercalciuria, is caused by mutations in the Na-K-2Cl cotransporter NKCC2. Nat Genet. 1996;13:183–8.
Simon DB, Karet FE, Rodriguez-Soriano J, et al. Genetic heterogeneity of Bartter’s syndrome revealed by mutations in the K+ channel, ROMK. Nat Genet. 1996;14:152–6.
Greger R. Ion transport mechanisms in thick ascending limb of Henle’s loop of mammalian nephron. Physiol Rev. 1985;65:760–97.
Seyberth H, Soergel M, Koeckerling A. Hypokalaemic tubular disorders: the hyperprostaglandin E syndrome and Gitelman-Bartter syndrome. In: Davison A, Cameron J, Grunfeld J, Kerr D, Ritz E, Winearls C, editors. Oxford textbook of clinical nephrology. Oxford: Oxford University Press; 1998. p. 1085–93.
Simon DB, Bindra RS, Mansfield TA, et al. Mutations in the chloride channel gene, CLCNKB, cause Bartter’s syndrome type III. Nat Genet. 1997;17:171–8.
Birkenhager R, Otto E, Schurmann MJ, et al. Mutation of BSND causes Bartter syndrome with sensorineural deafness and kidney failure. Nat Genet. 2001;29:310–4.
Estevez R, Boettger T, Stein V, et al. Barttin is a Cl- channel betasubunit crucial for renal Cl- reabsorption and inner ear K+ secretion. Nature. 2001;414:558–61.
Riccardi D, Brown EM. Physiology and pathophysiology of the calcium-sensing receptor in the kidney. Am J Physiol Renal Physiol. 2010;298:F485–99. This article is a recent review of the Ca ++ sensing receptor (CaSR) in the kidney.
Pearce SH, Williamson C, Kifor O, et al. A familial syndrome of hypocalcemia with hypercalciuria due to mutations in the calcium-sensing receptor. N Engl J Med. 1996;335:1115–22.
Pollak MR, Brown EM, Estep HL, et al. Autosomal dominant hypocalcaemia caused by a Ca(2+)-sensing receptor gene mutation. Nat Genet. 1994;8:303–7.
Watanabe S, Fukumoto S, Chang H, et al. Association between activating mutations of calcium-sensing receptor and Bartter’s syndrome. Lancet. 2002;360:692–4.
Vargas-Poussou R, Huang C, Hulin P, et al. Functional characterization of a calcium-sensing receptor mutation in severe autosomal dominant hypocalcemia with a Bartter-like syndrome. J Am Soc Nephrol. 2002;13:2259–66.
Wang W, Lu M, Balazy M, et al. Phospholipase A2 is involved in mediating the effect of extracellular Ca2+ on apical K+ channels in rat TAL. Am J Physiol. 1997;273:F421–9.
Simon DB, Lu Y, Choate KA, et al. Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science. 1999;285:103–6.
Konrad M, Schaller A, Seelow D, et al. Mutations in the tight junction gene claudin 19 (CLDN19) are associated with renal magnesium wasting, renal failure, and severe ocular involvement. Am J Hum Genet. 2006;79:949–57.
Praga M, Vara J, Gonzalez-Parra, et al. Familial hypomagnesemia with hypercalciuria and nephrocalcinosis. Kidney Int. 1995;47:1419–25.
Weber S, Schneider L, Peters M, et al. Novel paracellin-1 mutations in 25 families with familial hypomagnesemia with hypercalciuria and nephrocalcinosis. J Am Soc Nephrol. 2001;12:1872–81.
Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–8.
Wilcox ER, Burton QL, Naz S, et al. Mutations in the gene encoding tight junction claudin-14 cause autosomal recessive deafness DFNB29. Cell. 2001;104:165–72.
Ben-Yosef T, Belyantseva IA, Saunders TL, et al. Claudin 14 knockout mice, a model for autosomal recessive deafness DFNB29, are deaf due to cochlear hair cell degeneration. Hum Mol Genet. 2003;12:2049–61.
Elkouby-Naor L, Abassi Z, Lagziel A, et al. Double gene deletion reveals lack of cooperation between claudin 11 and claudin 14 tight junction proteins. Cell Tissue Res. 2008;333:427–38.
Kiuchi-Saishin Y, Gotoh S, Furuse M, et al. Differential expression patterns of claudins, tight junction membrane proteins, in mouse nephron segments. J Am Soc Nephrol. 2002;13:875–86.
Gong Y, Renigunta V, Himmerkus N, et al. Claudin-14 regulates renal Ca++ transport in response to CaSR signalling via a novel microRNA pathway. EMBO J. 2012;31(8):1999–2012. This article reveals the renal role of claudin-14 in Ca ++ metabolism and uncovers a novel signaling pathway for CaSR that utilizes the microRNA molecules.
Loupy A, Ramakrishnan SK, Wootla B, et al. PTH-independent regulation of blood calcium concentration by the calcium-sensing receptor. J Clin Invest. 2012;122(9):3355–67. This article reveals the renal function of CaSR independent of its role in the parathyroid glands.
Parks JH, Coward M, Coe FL. Correspondence between stone composition and urine supersaturation in nephrolithiasis. Kidney Int. 1997;51:894–900.
Evan AP, et al. Randall’s plaque of patients with nephrolithiasis begins in basement membranes of thin loops of Henle. J Clin Invest. 2003;111:607–16.
Kuo RL, et al. Urine calcium and volume predict coverage of renal papilla by Randall’s plaque. Kidney Int. 2003;64:2150–4.
Breiderhoff T, Himmerkus N, Stuiver M, et al. Deletion of claudin-10 (Cldn10) in the thick ascending limb impairs paracellular sodium permeability and leads to hypermagnesemia and nephrocalcinosis. Proc Natl Acad Sci U S A. 2012;109(35):14241–6. This article reports the renal phenotypes of claudin-10 knockout mice and reveals unexpected nephrocalcinosis accompanied by tubular hyperabsorption of Ca ++.
Vezzoli G, Terranegra A, Arcidiacono T, et al. R990G polymorphism of calcium-sensing receptor does produce a gain-of-function and predispose to primary hypercalciuria. Kidney Int. 2007;71:1155–62.
Corbetta S, Eller-Vainicher C, Filopanti M, et al. R990G polymorphism of the calcium-sensing receptor and renal calcium excretion in patients with primary hyperparathyroidism. Eur J Endocrinol. 2006;155:687–92.
Vezzoli G, Terranegra A, Arcidiacono T, et al. Calcium kidney stones are associated with a haplotype of the calcium-sensing receptor gene regulatory region. Nephrol Dial Transplant. 2010;25:2245–52.
Aloia A, Terranegra, A, Vezzoli G et al. Effects of the calcium sensing receptor promoter region polymorphisms in kidney stone disease. ASN 2011; [FR-PO1182].
Acknowledgements
This work was supported by the National Institutes of Health Grants RO1DK084059 and P30 DK079333, and American Heart Association Grant 0930050N.
Disclosure
The author reported no potential conflicts of interest relevant to this article.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hou, J. The Role of Claudin in Hypercalciuric Nephrolithiasis. Curr Urol Rep 14, 5–12 (2013). https://doi.org/10.1007/s11934-012-0289-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11934-012-0289-2