Abstract
Purpose of Review
Personalized medicine is a challenge to improve survival and quality of life of patients suffering from primary malignant brain tumor. Molecular biology is integrated in initial diagnosis and relapse, and, in the nearest future, over treatment schedule and monitoring. Liquid biopsy is a minimally invasive way to obtain tumor material.
Recent Findings
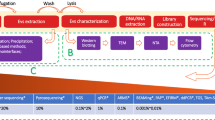
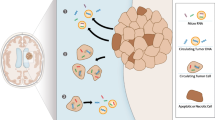
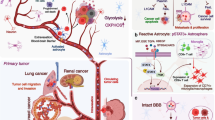
Over the past years, three fluids have been explored to provide tumor information in primary malignant brain tumor: blood, cerebrospinal fluid, and vitreous liquid. Different tumor components were identified: (1) circulating tumor cells, (2) circulating tumor DNA, (3) RNA and non-coding miRNA, and (4) extracellular vesicles. The performance of the liquid biopsy depends on the tumor type and on the method of detection.
Summary
Liquid biopsy could be a valuable tool to improve patient care in primary malignant brain tumor. Improvement of its sensitivity is the major challenge to generalize its use in daily practice.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance
de Robles P, Fiest KM, Frolkis AD, Pringsheim T, Atta C, St Germaine-Smith C, et al. The worldwide incidence and prevalence of primary brain tumors: a systematic review and meta-analysis. Neuro-Oncology. 2015;17(6):776–83. https://doi.org/10.1093/neuonc/nou283.
Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–20. https://doi.org/10.1007/s00401-016-1545-1.
Hodges TR, Ott M, Xiu J, Gatalica Z, Swensen J, Zhou S, et al. Mutational burden, immune checkpoint expression, and mismatch repair in glioma: implications for immune checkpoint immunotherapy. Neuro-Oncology. 2017;19(8):1047–57. https://doi.org/10.1093/neuonc/nox026.
Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16(11):2598–608. https://doi.org/10.1158/1535-7163.MCT-17-0386.
Le DT UJN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–20. https://doi.org/10.1056/NEJMoa1500596.
Reardon DA, Lassman AB, van den Bent M, Kumthekar P, Merrell R, Scott AM, et al. Efficacy and safety results of ABT-414 in combination with radiation and temozolomide in newly diagnosed glioblastoma. Neuro-Oncology. 2017;19(7):965–75. https://doi.org/10.1093/neuonc/now257.
Gan HK, Reardon DA, Lassman AB, Merrell R, van den Bent M, Butowski N, et al. Safety, pharmacokinetics and antitumor response of depatuxizumab mafodotin as monotherapy or in combination with temozolomide in patients with glioblastoma. Neuro-Oncology. 2017; https://doi.org/10.1093/neuonc/nox202.
Wilson WH, Young RM, Schmitz R, Yang Y, Pittaluga S, Wright G, et al. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nat Med. 2015;21(8):922–6. https://doi.org/10.1038/nm.3884.
Grommes C, Pastore A, Palaskas N, Tang SS, Campos C, Schartz D, et al. Ibrutinib unmasks critical role of Bruton tyrosine kinase in primary CNS lymphoma. Cancer Discov. 2017;7(9):1018–29. https://doi.org/10.1158/2159-8290.CD-17-0613.
Mohammad F, Weissmann S, Leblanc B, Pandey DP, Højfeldt JW, Comet I, et al. EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat Med. 2017;23(4):483–92. https://doi.org/10.1038/nm.4293.
Pi C, Zhang M-F, Peng X-X, Zhang Y-C, Xu C-R, Zhou Q. Liquid biopsy in non-small cell lung cancer: a key role in the future of personalized medicine? Expert Rev Mol Diagn. 2017;17(12):1089–96. https://doi.org/10.1080/14737159.2017.1395701.
De Mattos-Arruda L, Caldas C. Cell-free circulating tumour DNA as a liquid biopsy in breast cancer. Mol Oncol. 2016;10(3):464–74. https://doi.org/10.1016/j.molonc.2015.12.001.
Jia S, Zhang R, Li Z, Li J. Clinical and biological significance of circulating tumor cells, circulating tumor DNA, and exosomes as biomarkers in colorectal cancer. Oncotarget. 2017;8(33):55632–45. https://doi.org/10.18632/oncotarget.17184.
Masuda T, Hayashi N, Iguchi T, Ito S, Eguchi H, Mimori K. Clinical and biological significance of circulating tumor cells in cancer. Mol Oncol. 2016;10(3):408–17. https://doi.org/10.1016/j.molonc.2016.01.010.
Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10(20):6897–904. https://doi.org/10.1158/1078-0432.CCR-04-0378.
• Sullivan JP, Nahed BV, Madden MW, Oliveira SM, Springer S, Bhere D, et al. Brain tumor cells in circulation are enriched for mesenchymal gene expression. Cancer Discov. 2014;4:1299–309. This study provides evidence of release of tumor cells in blood in GBM and highlights that the release mechanism is dependent on a phenotype modification.
• Macarthur KM, Kao GD, Chandrasekaran S, Alonso-Basanta M, Chapman C, Lustig RA, et al. Detection of brain tumor cells in the peripheral blood by a telomerase promoter-based assay. Cancer Res. 2014;74:2152–9. This study develops an original method to detect circulating tumor cells in plasma.
• Schwaederle M, Chattopadhyay R, Kato S, Fanta PT, Banks KC, Choi IS, et al. Genomic alterations in circulating tumor DNA from diverse cancer patients identified by next-generation sequencing. Cancer Res. 2017;77:5419–27. This study highlights that a targeted NGS panel permits to detect somatic mutations and among them, targetable alterations, in plasma in PMBT.
Fontanilles M, Marguet F, Bohers É, Viailly P-J, Dubois S, Bertrand P, et al. Non-invasive detection of somatic mutations using next-generation sequencing in primary central nervous system lymphoma. Oncotarget. 2017;8(29):48157–68. https://doi.org/10.18632/oncotarget.18325.
Hattori K, Sakata-Yanagimoto M, Suehara Y, Yokoyama Y, Kato T, Kurita N, et al. Clinical significance of disease-specific MYD88 mutations in circulating DNA in primary central nervous system lymphoma. Cancer Sci. 2017;
Odjélé A, Charest D, Morin P. miRNAs as important drivers of glioblastomas: a no-brainer? Cancer Biomark. 2012;11(6):245–52. https://doi.org/10.3233/CBM-2012-0271.
• Manda SV, Kataria Y, Tatireddy BR, Ramakrishnan B, Ratnam BG, Lath R, et al. Exosomes as a biomarker platform for detecting epidermal growth factor receptor-positive high-grade gliomas. J. Neurosurg. 2017;1–11. This study highlights the use of exosomes in blood to identify high grade glioma using EGFR amplification.
• Figueroa JM, Skog J, Akers J, Li H, Komotar R, Jensen R, et al. Detection of wild-type EGFR amplification and EGFRvIII mutation in CSF-derived extracellular vesicles of glioblastoma patients. Neuro-Oncol. 2017;19:1494–502. This study is the first to report the two most common EGFR alterations in CSF.
• Huang TY, Piunti A, Lulla RR, Qi J, Horbinski CM, Tomita T, et al. Detection of Histone H3 mutations in cerebrospinal fluid-derived tumor DNA from children with diffuse midline glioma. Acta Neuropathol Commun. 2017;5:28. This study describes for the first time the detection of histone mutations H3 in CSF in children and opens the possibility to perform this method in midline gliomas in adults, which are particularly difficult to access surgically.
Akers JC, Hua W, Li H, Ramakrishnan V, Yang Z, Quan K, et al. A cerebrospinal fluid microRNA signature as biomarker for glioblastoma. Oncotarget. 2017;8(40):68769–79. https://doi.org/10.18632/oncotarget.18332.
Cani AK, Hovelson DH, Demirci H, Johnson MW, Tomlins SA, Rao RC. Next generation sequencing of vitreoretinal lymphomas from small-volume intraocular liquid biopsies: new routes to targeted therapies. Oncotarget. 2017;8(5):7989–98. https://doi.org/10.18632/oncotarget.14008.
Müller C, Holtschmidt J, Auer M, Heitzer E, Lamszus K, Schulte A, et al. Hematogenous dissemination of glioblastoma multiforme. Sci Transl Med. 2014;6(247):247ra101. https://doi.org/10.1126/scitranslmed.3009095.
Stroun M, Lyautey J, Lederrey C, Olson-Sand A, Anker P. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin Chim Acta. 2001;313:139–42.
Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61(4):1659–65.
Aucamp J, Bronkhorst AJ, Peters DL, Van Dyk HC, Van der Westhuizen FH, Pretorius PJ. Kinetic analysis, size profiling, and bioenergetic association of DNA released by selected cell lines in vitro. Cell Mol Life Sci. 2017;74(14):2689–707. https://doi.org/10.1007/s00018-017-2495-z.
Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985–90. https://doi.org/10.1038/nm.1789.
Medina Diaz I, Nocon A, Mehnert DH, Fredebohm J, Diehl F, Holtrup F. Performance of Streck cfDNA blood collection tubes for liquid biopsy testing. PLoS One. 2016;11(11):e0166354. https://doi.org/10.1371/journal.pone.0166354.
Norton SE, Lechner JM, Williams T, Fernando MR. A stabilizing reagent prevents cell-free DNA contamination by cellular DNA in plasma during blood sample storage and shipping as determined by digital PCR. Clin Biochem. 2013;46(15):1561–5. https://doi.org/10.1016/j.clinbiochem.2013.06.002.
Steffensen KD, Madsen CV, Andersen RF, Waldstrøm M, Adimi P, Jakobsen A. Prognostic importance of cell-free DNA in chemotherapy resistant ovarian cancer treated with bevacizumab. Eur J Cancer. 2014;50(15):2611–8. https://doi.org/10.1016/j.ejca.2014.06.022.
El Messaoudi S, Mouliere F, Du Manoir S, Bascoul-Mollevi C, Gillet B, Nouaille M, et al. Circulating DNA as a strong multimarker prognostic tool for metastatic colorectal cancer patient management care. Clin Cancer Res. 2016;22(12):3067–77. https://doi.org/10.1158/1078-0432.CCR-15-0297.
Yanagita M, Redig AJ, Paweletz CP, Dahlberg SE, O’Connell A, Feeney N, et al. A prospective evaluation of circulating tumor cells and cell-free DNA in EGFR-mutant non-small cell lung cancer patients treated with erlotinib on a phase II trial. Clin Cancer Res. 2016;22(24):6010–20. https://doi.org/10.1158/1078-0432.CCR-16-0909.
Rossi G, Mu Z, Rademaker A, Austin L, Strickland KS, Lima Barros Costa R, et al. Cell-free DNA and circulating tumor cells: comprehensive liquid biopsy analysis in advanced breast cancer. Clin Cancer Res. 2017; https://doi.org/10.1158/1078-0432.CCR-17-2092.
• De Mattos-Arruda L, Mayor R, CKY N, Weigelt B, Martínez-Ricarte F, Torrejon D, et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun. 2015;6:8839. This study was the first to compare diagnostic performance between blood and CSF using NGS in PMBT.
Rothé F, Laes J-F, Lambrechts D, Smeets D, Vincent D, Maetens M, et al. Plasma circulating tumor DNA as an alternative to metastatic biopsies for mutational analysis in breast cancer. Ann Oncol. 2014;25(10):1959–65. https://doi.org/10.1093/annonc/mdu288.
Boisselier B, Gállego Pérez-Larraya J, Rossetto M, Labussière M, Ciccarino P, Marie Y, et al. Detection of IDH1 mutation in the plasma of patients with glioma. Neurology. 2012;79:1693–8.
Fiano V, Trevisan M, Trevisan E, Senetta R, Castiglione A, Sacerdote C, et al. MGMT promoter methylation in plasma of glioma patients receiving temozolomide. J Neuro-Oncol. 2014;117(2):347–57. https://doi.org/10.1007/s11060-014-1395-4.
Majchrzak-Celińska A, Paluszczak J, Kleszcz R, Magiera M, Barciszewska A-M, Nowak S, et al. Detection of MGMT, RASSF1A, p15INK4B, and p14ARF promoter methylation in circulating tumor-derived DNA of central nervous system cancer patients. J Appl Genet. 2013;54(3):335–44. https://doi.org/10.1007/s13353-013-0149-x.
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–8. https://doi.org/10.1038/nature03702.
Li R, Gao K, Luo H, Wang X, Shi Y, Dong Q, et al. Identification of intrinsic subtype-specific prognostic microRNAs in primary glioblastoma. J Exp Clin Cancer Res. 2014;33(1):9. https://doi.org/10.1186/1756-9966-33-9.
Wei X, Chen D, Lv T, Li G, Qu S. Serum MicroRNA-125b as a potential biomarker for glioma diagnosis. Mol Neurobiol. 2016;53(1):163–70. https://doi.org/10.1007/s12035-014-8993-1.
Regazzo G, Terrenato I, Spagnuolo M, Carosi M, Cognetti G, Cicchillitti L, et al. A restricted signature of serum miRNAs distinguishes glioblastoma from lower grade gliomas. J Exp Clin Cancer Res. 2016;35(1):124. https://doi.org/10.1186/s13046-016-0393-0.
Wang Z-Q, Zhang M-Y, Deng M-L, Weng N-Q, Wang H-Y, Wu S-X. Low serum level of miR-485-3p predicts poor survival in patients with glioblastoma. PLoS One. 2017;12(9):e0184969. https://doi.org/10.1371/journal.pone.0184969.
Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–83. https://doi.org/10.1083/jcb.201211138.
Ruivo CF, Adem B, Silva M, Melo SA. The biology of cancer exosomes: insights and new perspectives. Cancer Res. 2017;
Kucharzewska P, Christianson HC, Welch JE, Svensson KJ, Fredlund E, Ringnér M, et al. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc Natl Acad Sci U S A. 2013;110(18):7312–7. https://doi.org/10.1073/pnas.1220998110.
Manterola L, Guruceaga E, Gállego Pérez-Larraya J, González-Huarriz M, Jauregui P, Tejada S, et al. A small noncoding RNA signature found in exosomes of GBM patient serum as a diagnostic tool. Neuro-Oncology. 2014;16(4):520–7. https://doi.org/10.1093/neuonc/not218.
Chen WW, Balaj L, Liau LM, Samuels ML, Kotsopoulos SK, Maguire CA, et al. BEAMing and droplet digital PCR analysis of mutant IDH1 mRNA in glioma patient serum and cerebrospinal fluid extracellular vesicles. Mol Ther Nucleic Acids. 2013;2:e109. https://doi.org/10.1038/mtna.2013.28.
Pan W, Gu W, Nagpal S, Gephart MH, Quake SR. Brain tumor mutations detected in cerebral spinal fluid. Clin Chem. 2015;61(3):514–22. https://doi.org/10.1373/clinchem.2014.235457.
Baraniskin A, Kuhnhenn J, Schlegel U, Schmiegel W, Hahn S, Schroers R. MicroRNAs in cerebrospinal fluid as biomarker for disease course monitoring in primary central nervous system lymphoma. J Neuro-Oncol. 2012;109(2):239–44. https://doi.org/10.1007/s11060-012-0908-2.
Baraniskin A, Zaslavska E, Nöpel-Dünnebacke S, Ahle G, Seidel S, Schlegel U, et al. Circulating U2 small nuclear RNA fragments as a novel diagnostic biomarker for primary central nervous system lymphoma. Neuro-Oncology. 2015;
Drusco A, Bottoni A, Laganà A, Acunzo M, Fassan M, Cascione L, et al. A differentially expressed set of microRNAs in cerebro-spinal fluid (CSF) can diagnose CNS malignancies. Oncotarget. 2015;6(25):20829–39. https://doi.org/10.18632/oncotarget.4096.
Pochat-Cotilloux C, Bienvenu J, Nguyen A-M, Ohanessian R, Ghesquières H, Sève P, et al. Use of a threshold of interleukin-10 and IL-10/IL-6 ratio in ocular samples for the screening of vitreoretinal lymphoma. Retina (Philadelphia, Pa). 2017;
Bonzheim I, Giese S, Deuter C, Süsskind D, Zierhut M, Waizel M, et al. High frequency of MYD88 mutations in vitreoretinal B-cell lymphoma: a valuable tool to improve diagnostic yield of vitreous aspirates. Blood. 2015;126(1):76–9. https://doi.org/10.1182/blood-2015-01-620518.
Noel N, Couteau J, Maillet G, Gobet F, D’Aloisio F, Minier C, et al. TP53 and FGFR3 gene mutation assessment in urine: pilot study for bladder cancer diagnosis. Anticancer Res. 2015;35(9):4915–21.
Kinde I, Munari E, Faraj SF, Hruban RH, Schoenberg M, Bivalacqua T, et al. TERT promoter mutations occur early in urothelial neoplasia and are biomarkers of early disease and disease recurrence in urine. Cancer Res. 2013;73(24):7162–7. https://doi.org/10.1158/0008-5472.CAN-13-2498.
Botezatu I, Serdyuk O, Potapova G, Shelepov V, Alechina R, Molyaka Y, et al. Genetic analysis of DNA excreted in urine: a new approach for detecting specific genomic DNA sequences from cells dying in an organism. Clin Chem. 2000;46(8 Pt 1):1078–84.
Chen S, Zhao J, Cui L, Liu Y. Urinary circulating DNA detection for dynamic tracking of EGFR mutations for NSCLC patients treated with EGFR-TKIs. Clin Transl Oncol. 2017;19(3):332–40. https://doi.org/10.1007/s12094-016-1534-9.
Reckamp KL, Melnikova VO, Karlovich C, Sequist LV, Camidge DR, Wakelee H, et al. A highly sensitive and quantitative test platform for detection of NSCLC EGFR mutations in urine and plasma. J Thorac Oncol. 2016;11(10):1690–700. https://doi.org/10.1016/j.jtho.2016.05.035.
Pu D, Liang H, Wei F, Akin D, Feng Z, Yan Q, et al. Evaluation of a novel saliva-based epidermal growth factor receptor mutation detection for lung cancer: a pilot study. Thorac Cancer. 2016;7(4):428–36. https://doi.org/10.1111/1759-7714.12350.
Jamal-Hanjani M, Wilson GA, Horswell S, Mitter R, Sakarya O, Constantin T, et al. Detection of ubiquitous and heterogeneous mutations in cell-free DNA from patients with early-stage non-small-cell lung cancer. Ann Oncol. 2016;27(5):862–7. https://doi.org/10.1093/annonc/mdw037.
Hohaus S, Giachelia M, Massini G, Mansueto G, Vannata B, Bozzoli V, et al. Cell-free circulating DNA in Hodgkin’s and non-Hodgkin’s lymphomas. Ann Oncol. 2009;20(8):1408–13. https://doi.org/10.1093/annonc/mdp006.
• Underhill HR, Kitzman JO, Hellwig S, Welker NC, Daza R, Baker DN, et al. Fragment length of circulating tumor DNA. PLoS Genet. 2016;12:e1006162. This study explores the mechanism of release of ctDNA in plasma in GBM and provides tools to improve detection method in future studies.
Madhavan D, Wallwiener M, Bents K, Zucknick M, Nees J, Schott S, et al. Plasma DNA integrity as a biomarker for primary and metastatic breast cancer and potential marker for early diagnosis. Breast Cancer Res Treat. 2014;146(1):163–74. https://doi.org/10.1007/s10549-014-2946-2.
Leng S, Zheng J, Jin Y, Zhang H, Zhu Y, Wu J, et al. Plasma cell-free DNA level and its integrity as biomarkers to distinguish non-small cell lung cancer from tuberculosis. Clin Chim Acta. 2017;
Best MG, Sol N, In ‘t Veld SGJG, Vancura A, Muller M, A-LN N, et al. Swarm intelligence-enhanced detection of non-small-cell lung cancer using tumor-educated platelets. Cancer Cell. 2017;32:238–252.e9.
Krug AK, Enderle D, Karlovich C, Priewasser T, Bentink S, Spiel A, et al. Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Ann Oncol. 2017; https://doi.org/10.1093/annonc/mdx765.
Uehiro N, Sato F, Pu F, Tanaka S, Kawashima M, Kawaguchi K, et al. Circulating cell-free DNA-based epigenetic assay can detect early breast cancer. Breast Cancer Res. 2016;18(1):129. https://doi.org/10.1186/s13058-016-0788-z.
Acknowledgements
The authors thank Louise Damian for providing graphical support for figure elaboration. The research leading to these results has received funding from the program “Investissements d’Avenir” ANR-10-IAIHU-06, Institut Universitaire de Cancérologie.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
A.D.-P. declares no conflict of interest.
M.F. reports non-financial support from Hoffman-La Roche and Amgen and personal fees from “La Lettre du Cancérologue,” outside the submitted work. A.I. reports grants from Fondation ARC, other from IntselChimos, other from Hoffman-La Roche, other from Beta-Innov (July 2014); personal fees from “La Lettre du Cancérologue,” other from Cathera (June 2017), other from BMS (November 2015), other from Hoffman-La Roche (December 2015), other from Cipla (December 2015), outside the submitted work.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical collection on Neuro-oncology
Rights and permissions
About this article
Cite this article
Fontanilles, M., Duran-Peña, A. & Idbaih, A. Liquid Biopsy in Primary Brain Tumors: Looking for Stardust!. Curr Neurol Neurosci Rep 18, 13 (2018). https://doi.org/10.1007/s11910-018-0820-z
Published:
DOI: https://doi.org/10.1007/s11910-018-0820-z