Abstract
Purpose of Review
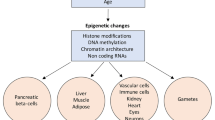
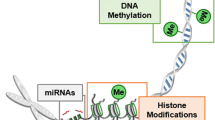
Great strides have recently been made in elucidating the role of genetic sequence variation in diabetes pathogenesis. Increasingly, studies are focusing on other factors that may contribute to the pathogenesis of diabetes, such as epigenetics, a term “traditionally” encompassing changes to the DNA that do not alter sequence and are heritable (primary methylation and histone modification) but often expanded to include microRNAs. This review summarizes latest findings on the role of epigenetics in diabetes pathogenesis.
Recent Findings
Recent studies illustrate roles for methylation changes, histone modification, imprinting, and microRNAs across several diabetes types and complications. Notably, methylation changes in the human leukocyte antigen (HLA) region have been found to precede the development of type 1 diabetes. In type 2 diabetes, lifestyle factors appear to interact with epigenetic mechanisms in pathogenesis.
Summary
Emerging technologies have allowed increasingly comprehensive descriptive analysis of the role of epigenetic mechanisms in diabetes pathogenesis which have yielded meaningful insights into effects on expression of relevant genes. These findings have the potential to inform future development of predictive testing to enable primary prevention and further work to uncover the complex pathogenesis of diabetes.
Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major Importance
Rowley WR, Bezold C, Arikan Y, Byrne E, Krohe S. Diabetes 2030: insights from yesterday, today, and future trends. Popul Health Manag. 2017;20:6–12.
Groop L, Pociot F. Genetics of diabetes--are we missing the genes or the disease? Mol Cell Endocrinol. 2014;382:726–39.
Pociot F, Lernmark A. Genetic risk factors for type 1 diabetes. Lancet. 2016;387:2331–9.
Fuchsberger C, Flannick J, Teslovich TM, Mahajan A, Agarwala V, Gaulton KJ, et al. The genetic architecture of type 2 diabetes. Nature. 2016;536:41–7.
Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–32.
Aziz A, Liu QC, Dilworth FJ. Regulating a master regulator: establishing tissue-specific gene expression in skeletal muscle. Epigenetics. 2010;5:691–5.
Rakyan VK, Beyan H, Down TA, Hawa MI, Maslau S, Aden D, et al. Identification of type 1 diabetes-associated DNA methylation variable positions that precede disease diagnosis. PLoS Genet. 2011;7:e1002300.
Stankov K, Benc D, Draskovic D. Genetic and epigenetic factors in etiology of diabetes mellitus type 1. Pediatrics. 2013;132:1112–22.
Katada S, Imhof A, Sassone-Corsi P. Connecting threads: epigenetics and metabolism. Cell. 2012;148:24–8.
Wang Z, Xie Z, Lu Q, Chang C, Zhou Z. Beyond genetics: What causes type 1 diabetes. Clin Rev Allergy Immunol. 2017;52:273–86.
Drong AW, Lindgren CM, McCarthy MI. The genetic and epigenetic basis of type 2 diabetes and obesity. Clin Pharmacol Ther. 2012;92:707–15.
Stefan M, Zhang W, Concepcion E, Yi Z, Tomer Y. DNA methylation profiles in type 1 diabetes twins point to strong epigenetic effects on etiology. J Autoimmun. 2014;50:33–7.
Bell GI, Horita S, Karam JH. A polymorphic locus near the human insulin gene is associated with insulin-dependent diabetes mellitus. Diabetes. 1984;33:176–83.
Rui J, Deng S, Lebastchi J, Clark PL, Usmani-Brown S, Herold KC. Methylation of insulin DNA in response to proinflammatory cytokines during the progression of autoimmune diabetes in NOD mice. Diabetologia. 2016;59:1021–9.
Akirav EM, Lebastchi J, Galvan EM, Henegariu O, Akirav M, Ablamunits V, et al. Detection of beta cell death in diabetes using differentially methylated circulating DNA. Proc Natl Acad Sci U S A. 2011;108:19018–23.
Husseiny MI, Kuroda A, Kaye AN, Nair I, Kandeel F, Ferreri K. Development of a quantitative methylation-specific polymerase chain reaction method for monitoring beta cell death in type 1 diabetes. PLoS One. 2012;7:e47942.
Fisher MM, Watkins RA, Blum J, Evans-Molina C, Chalasani N, DiMeglio LA, et al. Elevations in circulating methylated and unmethylated preproinsulin DNA in new-onset type 1 diabetes. Diabetes. 2015;64:3867–72.
Fradin D, Le Fur S, Mille C, Naoui N, Groves C, Zelenika D, et al. Association of the CpG methylation pattern of the proximal insulin gene promoter with type 1 diabetes. PLoS One. 2012;7:e36278.
•• Volkov P, Bacos K, Ofori JK, Esguerra JL, Eliasson L, Ronn T, et al. Whole-genome bisulfite sequencing of human pancreatic islets reveals novel differentially methylated regions in type 2 diabetes pathogenesis. Diabetes. 2017;66:1074–85. Comprehensive whole-genome bisulfite sequencing analysis of DNA methylation of human islet cells from a T2D case/control study.
Szabat M, Kalynyak TB, Lim GE, Chu KY, Yang YH, Asadi A, et al. Musashi expression in beta-cells coordinates insulin expression, apoptosis and proliferation in response to endoplasmic reticulum stress in diabetes. Cell Death Dis. 2011;2:e232.
Jeon JP, Koh IU, Choi NH, Kim BJ, Han BG, Lee S. Differential DNA methylation of MSI2 and its correlation with diabetic traits. PLoS One. 2017;12:e0177406.
• Dhana K, Braun KVE, Nano J, Voortman T, Demerath EW, Guan W, Fornage M, van Meurs JBJ, Uitterlinden AG, Hofman A, Franco OH, and Dehghan A. An epigenome-wide association study (EWAS) of obesity-related traits. Am J Epidemiol. 2018. This study replicates previous results from African American on obesity-related traits, and also found novel association between DNA methylation and increased BMI in older adults.
You D, Nilsson E, Tenen DE, Lyubetskaya A, Lo JC, Jiang R, Deng J, Dawes BA, Vaag A, Ling C, Rosen ED, and Kang S. Dnmt3a is an epigenetic mediator of adipose insulin resistance. Elife. 2017; 6. https://doi.org/10.7554/eLife.30766.
Dayeh T, Tuomi T, Almgren P, Perfilyev A, Jansson PA, de Mello VD, et al. DNA methylation of loci within ABCG1 and PHOSPHO1 in blood DNA is associated with future type 2 diabetes risk. Epigenetics. 2016;11:482–8.
Hall E, Dekker Nitert M, Volkov P, Malmgren S, Mulder H, Bacos K, et al. The effects of high glucose exposure on global gene expression and DNA methylation in human pancreatic islets. Mol Cell Endocrinol. 2018;472:57–67.
Gillberg L, Perfilyev A, Brons C, Thomasen M, Grunnet LG, Volkov P, et al. Adipose tissue transcriptomics and epigenomics in low birthweight men and controls: role of high-fat overfeeding. Diabetologia. 2016;59:799–812.
Liang H, Ward WF. PGC-1alpha: a key regulator of energy metabolism. Adv Physiol Educ. 2006;30:145–51.
Barres R, Osler ME, Yan J, Rune A, Fritz T, Caidahl K, et al. Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab. 2009;10:189–98.
Henagan TM, Stefanska B, Fang Z, Navard AM, Ye J, Lenard NR, et al. Sodium butyrate epigenetically modulates high-fat diet-induced skeletal muscle mitochondrial adaptation, obesity and insulin resistance through nucleosome positioning. Br J Pharmacol. 2015;172:2782–98.
Bajpeyi S, Covington JD, Taylor EM, Stewart LK, Galgani JE, Henagan TM. Skeletal muscle PGC1alpha −1 nucleosome position and −260 nt DNA methylation determine exercise response and prevent ectopic lipid accumulation in men. Endocrinology. 2017;158:2190–9.
Elhamamsy AR. Role of DNA methylation in imprinting disorders: an updated review. J Assist Reprod Genet. 2017;34:549–62.
Docherty LE, Kabwama S, Lehmann A, Hawke E, Harrison L, Flanagan SE, et al. Clinical presentation of 6q24 transient neonatal diabetes mellitus (6q24 TNDM) and genotype-phenotype correlation in an international cohort of patients. Diabetologia. 2013;56:758–62.
Hoffmann A, Spengler D. Role of ZAC1 in transient neonatal diabetes mellitus and glucose metabolism. World J Biol Chem. 2015;6:95–109.
Wallace C, Smyth DJ, Maisuria-Armer M, Walker NM, Todd JA, Clayton DG. The imprinted DLK1-MEG3 gene region on chromosome 14q32.2 alters susceptibility to type 1 diabetes. Nat Genet. 2010;42:68–71.
Prats-Puig A, Carreras-Badosa G, Bassols J, Cavelier P, Magret A, Sabench C, et al. The placental imprinted DLK1-DIO3 domain: a new link to prenatal and postnatal growth in humans. Am J Obstet Gynecol. 2017;217:350.e1–350.e13.
Benetatos L, Hatzimichael E, Londin E, Vartholomatos G, Loher P, Rigoutsos I, et al. The microRNAs within the DLK1-DIO3 genomic region: involvement in disease pathogenesis. Cell Mol Life Sci. 2013;70:795–814.
Kameswaran V, Bramswig NC, McKenna LB, Penn M, Schug J, Hand NJ, et al. Epigenetic regulation of the DLK1-MEG3 microRNA cluster in human type 2 diabetic islets. Cell Metab. 2014;19:135–45.
Small KS, Hedman AK, Grundberg E, Nica AC, Thorleifsson G, Kong A, et al. Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat Genet. 2011;43:561–4.
Miao F, Chen Z, Zhang L, Liu Z, Wu X, Yuan YC, et al. Profiles of epigenetic histone post-translational modifications at type 1 diabetes susceptible genes. J Biol Chem. 2012;287:16335–45.
Nistico L, Buzzetti R, Pritchard LE, Van der Auwera B, Giovannini C, Bosi E, et al. The CTLA-4 gene region of chromosome 2q33 is linked to, and associated with, type 1 diabetes. Belgian diabetes registry. Hum Mol Genet. 1996;5:1075–80.
Miao F, Smith DD, Zhang L, Min A, Feng W, Natarajan R. Lymphocytes from patients with type 1 diabetes display a distinct profile of chromatin histone H3 lysine 9 dimethylation: an epigenetic study in diabetes. Diabetes. 2008;57:3189–98.
Li Y, Reddy MA, Miao F, Shanmugam N, Yee JK, Hawkins D, et al. Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-kappaB-dependent inflammatory genes. Relevance to diabetes and inflammation. J Biol Chem. 2008;283:26771–81.
Paneni F, Costantino S, Battista R, Castello L, Capretti G, Chiandotto S, et al. Adverse epigenetic signatures by histone methyltransferase Set7 contribute to vascular dysfunction in patients with type 2 diabetes mellitus. Circ Cardiovasc Genet. 2015;8:150–8.
Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97.
Tammen SA, Friso S, Choi SW. Epigenetics: the link between nature and nurture. Mol Asp Med. 2013;34:753–64.
Wang G, Gu Y, Xu N, Zhang M, Yang T. Decreased expression of miR-150, miR146a and miR424 in type 1 diabetic patients: Association with ongoing islet autoimmunity. Biochem Biophys Res Commun. 2018;498:382–7.
Assmann TS, Duarte GC, Brondani LA, de Freitas PH, Martins EM, Canani LH, et al. Polymorphisms in genes encoding miR-155 and miR-146a are associated with protection to type 1 diabetes mellitus. Acta Diabetol. 2017;54:433–41.
Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–14.
Erener S, Mojibian M, Fox JK, Denroche HC, Kieffer TJ. Circulating miR-375 as a biomarker of beta-cell death and diabetes in mice. Endocrinology. 2013;154:603–8.
Song I, Roels S, Martens GA, Bouwens L. Circulating microRNA-375 as biomarker of pancreatic beta cell death and protection of beta cell mass by cytoprotective compounds. PLoS One. 2017;12:e0186480.
Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–74.
Barutta F, Bruno G, Matullo G, Chaturvedi N, Grimaldi S, Schalkwijk C, et al. MicroRNA-126 and micro−/macrovascular complications of type 1 diabetes in the EURODIAB prospective complications study. Acta Diabetol. 2017;54:133–9.
Zampetaki A, Willeit P, Burr S, Yin X, Langley SR, Kiechl S, et al. Angiogenic microRNAs linked to incidence and progression of diabetic retinopathy in type 1 diabetes. Diabetes. 2016;65:216–27.
Satake E, Pezzolesi MG, Md Dom ZI, Smiles AM, Niewczas MA, Krolewski AS. Circulating miRNA profiles associated with hyperglycemia in patients with type 1 diabetes. Diabetes. 2018;67:1013–23.
He Y, Ding Y, Liang B, Lin J, Kim TK, Yu H, Hang H, and Wang K. A systematic study of dysregulated MicroRNA in type 2 diabetes mellitus. Int J Mol Sci. 2017;18. https://doi.org/10.3390/ijms18030456.
Bao L, Fu X, Si M, Wang Y, Ma R, Ren X, et al. MicroRNA-185 targets SOCS3 to inhibit beta-cell dysfunction in diabetes. PLoS One. 2015;10:e0116067.
Li Y, Luo T, Wang L, Wu J, Guo S. MicroRNA-19a-3p enhances the proliferation and insulin secretion, while it inhibits the apoptosis of pancreatic beta cells via the inhibition of SOCS3. Int J Mol Med. 2016;38:1515–24.
Wu H, Zhang T, Pan F, Steer CJ, Li Z, Chen X, et al. MicroRNA-206 prevents hepatosteatosis and hyperglycemia by facilitating insulin signaling and impairing lipogenesis. J Hepatol. 2017;66:816–24.
Yang WM, Min KH, Lee W. Induction of miR-96 by dietary saturated fatty acids exacerbates hepatic insulin resistance through the suppression of INSR and IRS-1. PLoS One. 2016;11:e0169039.
Massart J, Sjogren RJO, Lundell LS, Mudry JM, Franck N, O'Gorman DJ, et al. Altered miR-29 expression in type 2 diabetes influences glucose and lipid metabolism in skeletal muscle. Diabetes. 2017;66:1807–18.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Haichen Zhang has no conflicts to declare.
Toni I. Pollin reports grant support from Regeneron Genetics Center.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Genetics
Rights and permissions
About this article
Cite this article
Zhang, H., Pollin, T.I. Epigenetics Variation and Pathogenesis in Diabetes. Curr Diab Rep 18, 121 (2018). https://doi.org/10.1007/s11892-018-1091-4
Published:
DOI: https://doi.org/10.1007/s11892-018-1091-4