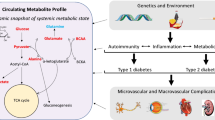
Abstract
Type 2 diabetes (T2D) is increasing worldwide, making identification of biomarkers for detection, staging, and effective prevention strategies an especially critical scientific and medical goal. Fortunately, advances in metabolomics techniques, together with improvements in bioinformatics and mathematical modeling approaches, have provided the scientific community with new tools to describe the T2D metabolome. The metabolomics signatures associated with T2D and obesity include increased levels of lactate, glycolytic intermediates, branched-chain and aromatic amino acids, and long-chain fatty acids. Conversely, tricarboxylic acid cycle intermediates, betaine, and other metabolites decrease. Future studies will be required to fully integrate these and other findings into our understanding of diabetes pathophysiology and to identify biomarkers of disease risk, stage, and responsiveness to specific treatments.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance,•• Of major importance
DeFronzo RA. Banting lecture. From the triumvirate to the ominous octet: a new paradigm for the treatment of type 2 diabetes mellitus. Diabetes. 2009;58(4):773–95.
Tabak AG, Jokela M, Akbaraly TN, Brunner EJ, Kivimaki M, Witte DR. Trajectories of glycaemia, insulin sensitivity, and insulin secretion before diagnosis of type 2 diabetes: an analysis from the Whitehall II study. Lancet. 2009;373(9682):2215–21.
Martin BC, Warram JH, Rosner B, Rich SS, Soeldner JS, Krolewski AS. Familial clustering of insulin sensitivity. Diabetes. 1992;41:850–4.
IDF. IDF diabetes atlas. Seventh ed. Brussels, Belgium: International Diabetes Federation; 2015. p. 144.
Pauling L, Robinson AB, Teranishi R, Cary P. Quantitative analysis of urine vapor and breath by gas-liquid partition chromatography. Proc Natl Acad Sci U S A. 1971;68(10):2374–6.
Rhee EP, Gerszten RE. Metabolomics and cardiovascular biomarker discovery. Clin Chem. 2012;58(1):139–47.
Bain JR, Stevens RD, Wenner BR, Ilkayeva O, Muoio DM, Newgard CB. Metabolomics applied to diabetes research. Moving from information to knowledge. Diabetes. 2009;58:2429–43.
Patti GJ, Yanes O, Siuzdak G. Innovation: metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13(4):263–9.
Roberts LD, Gerszten RE. Toward new biomarkers of cardiometabolic diseases. Cell Metab. 2013;18(1):43–50.
Drogan D, Dunn WB, Lin W, Buijsse B, Schulze MB, Langenberg C, et al. Untargeted metabolic profiling identifies altered serum metabolites of type 2 diabetes mellitus in a prospective, nested case control study. Clin Chem. 2015;61(3):487–97.
Park S, Sadanala KC, Kim EK. A metabolomic approach to understanding the metabolic link between obesity and diabetes. Mol Cells. 2015;38(7):587–96.
Fiehn O, Garvey WT, Newman JW, Lok KH, Hoppel CL, Adams SH. Plasma metabolomic profiles reflective of glucose homeostasis in non-diabetic and type 2 diabetic obese African-American women. PLoS One. 2010;5(12):e15234.
Gogna N, Krishna M, Oommen AM, Dorai K. Investigating correlations in the altered metabolic profiles of obese and diabetic subjects in a South Indian Asian population using an NMR-based metabolomic approach. Mol Biosyst. 2015;11(2):595–606.
Xu F, Tavintharan S, Sum CF, Woon K, Lim SC, Ong CN. Metabolic signature shift in type 2 diabetes mellitus revealed by mass spectrometry-based metabolomics. J Clin Endocrinol Metab. 2013;98(6):E1060–E5.
Suhre K, Meisinger C, Doring A, Altmaier E, Belcredi P, Gieger C, et al. Metabolic footprint of diabetes: a multiplatform metabolomics study in an epidemiological setting. PLoS One. 2010;5(11):e13953.
Menni C, Fauman E, Erte I, Perry JR, Kastenmuller G, Shin SY, et al. Biomarkers for type 2 diabetes and impaired fasting glucose using a nontargeted metabolomics approach. Diabetes. 2013;62(12):4270–6.
Shaham O, Wei R, Wang TJ, Ricciardi C, Lewis GD, Vasan RS, et al. Metabolic profiling of the human response to a glucose challenge reveals distinct axes of insulin sensitivity. Mol Syst Biol. 2008;4:214.
Spegel P, Malmgren S, Sharoyko VV, Newsholme P, Koeck T, Mulder H. Metabolomic analyses reveal profound differences in glycolytic and tricarboxylic acid cycle metabolism in glucose-responsive and -unresponsive clonal beta-cell lines. Biochem J. 2011;435(1):277–84.
Boucher A, Lu D, Burgess SC, Telemaque-Potts S, Jensen MV, Mulder H, et al. Biochemical mechanism of lipid-induced impairment of glucose-stimulated insulin secretion and reversal with a malate analogue. J Biol Chem. 2004;279(26):27263–71.
Lu D, Mulder H, Zhao P, Burgess SC, Jensen MV, Kamzolova S, et al. 13C NMR isotopomer analysis reveals a connection between pyruvate cycling and glucose-stimulated insulin secretion (GSIS). Proc Natl Acad Sci U S A. 2002;99(5):2708–13.
Kelley DE, Mokan M, Simoneau JA, Mandarino LJ. Interaction between glucose and free fatty acid metabolism in human skeletal muscle. J Clin Invest. 1993;92(1):91–8.
Koves TR, Li P, An J, Akimoto T, Slentz D, Ilkayeva O, et al. Peroxisome proliferator-activated receptor-gamma co-activator 1alpha-mediated metabolic remodeling of skeletal myocytes mimics exercise training and reverses lipid-induced mitochondrial inefficiency. J Biol Chem. 2005;280(39):33588–98.
Koves TR, Ussher JR, Noland RC, Slentz D, Mosedale M, Ilkayeva O, et al. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 2008;7(1):45–56.
Giesbertz P, Padberg I, Rein D, Ecker J, Hofle AS, Spanier B, et al. Metabolite profiling in plasma and tissues of ob/ob and db/db mice identifies novel markers of obesity and type 2 diabetes. Diabetologia. 2015;58(9):2133–43.
Kowalski GM, De Souza DP, Burch ML, Hamley S, Kloehn J, Selathurai A, et al. Application of dynamic metabolomics to examine in vivo skeletal muscle glucose metabolism in the chronically high-fat fed mouse. Biochem Biophys Res Commun. 2015;462(1):27–32.
Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9(4):311–26.
Salek RM, Maguire ML, Bentley E, Rubtsov DV, Hough T, Cheeseman M, et al. A metabolomic comparison of urinary changes in type 2 diabetes in mouse, rat, and human. Physiol Genomics. 2007;29(2):99–108.
Connor SC, Hansen MK, Corner A, Smith RF, Ryan TE. Integration of metabolomics and transcriptomics data to aid biomarker discovery in type 2 diabetes. Mol Biosyst. 2010;6(5):909–21.
Adams SH, Hoppel CL, Lok KH, Zhao L, Wong SW, Minkler PE, et al. Plasma acylcarnitine profiles suggest incomplete long-chain fatty acid beta-oxidation and altered tricarboxylic acid cycle activity in type 2 diabetic African-American women. J Nutr. 2009;139(6):1073–81.
Huffman KM, Shah SH, Stevens RD, Bain JR, Muehlbauer M, Slentz CA, et al. Relationships between circulating metabolic intermediates and insulin action in overweight to obese, inactive men and women. Diabetes Care. 2009;32(9):1678–83.
Felig P, Marliss E, Cahill Jr GF. Plasma amino acid levels and insulin secretion in obesity. N Engl J Med. 1969;281(15):811–6.
Wurtz P, Soininen P, Kangas AJ, Ronnemaa T, Lehtimaki T, Kahonen M, et al. Branched-chain and aromatic amino acids are predictors of insulin resistance in young adults. Diabetes Care. 2013;36(3):648–55.
Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17(4):448–53. This study demonstrates how a combination of three amino acids predicts future diabetes, suggesting that amino acid profiles in patients might help in assessing disease risk in susceptible individuals.
Shin AC, Fasshauer M, Filatova N, Grundell LA, Zielinski E, Zhou JY, et al. Brain insulin lowers circulating BCAA levels by inducing hepatic BCAA catabolism. Cell Metab. 2014;20(5)):898–909. This study demonstrates how elevated BCAA levels in obesity and diabetes (which can predict T2D) are reduced after insulin treatment. Moreover, brain insulin signaling is shown as a major regulator of BCAA metabolism by inducing hepatic BCKDH (rate-limiting enzyme in BCAA degradation).
Yamakado M, Tanaka T, Nagao K, Ishizaka Y, Mitushima T, Tani M, et al. Plasma amino acid profile is associated with visceral fat accumulation in obese Japanese subjects. Clin Obes. 2012;2(1–2):29–40.
Gall WE, Beebe K, Lawton KA, Adam KP, Mitchell MW, Nakhle PJ, et al. alpha-hydroxybutyrate is an early biomarker of insulin resistance and glucose intolerance in a nondiabetic population. PLoS One. 2010;5(5):e10883.
Cheng S, Rhee EP, Larson MG, Lewis GD, McCabe EL, Shen D, et al. Metabolite profiling identifies pathways associated with metabolic risk in humans. Circulation. 2012;125(18):2222–31.
Wurtz P, Tiainen M, Makinen VP, Kangas AJ, Soininen P, Saltevo J, et al. Circulating metabolite predictors of glycemia in middle-aged men and women. Diabetes Care. 2012;35(8):1749–56.
Stancakova A, Civelek M, Saleem NK, Soininen P, Kangas AJ, Cederberg H, et al. Hyperglycemia and a common variant of GCKR are associated with the levels of eight amino acids in 9,369 Finnish men. Diabetes. 2012;61(7):1895–902.
Wang-Sattler R, Yu Z, Herder C, Messias AC, Floegel A, He Y, et al. Novel biomarkers for pre-diabetes identified by metabolomics. Mol Syst Biol. 2012;8:615.
Sekhar RV, McKay SV, Patel SG, Guthikonda AP, Reddy VT, Balasubramanyam A, et al. Glutathione synthesis is diminished in patients with uncontrolled diabetes and restored by dietary supplementation with cysteine and glycine. Diabetes Care. 2011;34(1):162–7.
Ejaz A, Martinez-Guino L, Goldfine AB, Ribas-Aulinas F, De Nigris V, Ribo S, et al. Dietary betaine supplementation increases Fgf21 levels to improve glucose homeostasis and reduce hepatic lipid accumulation in mice. Diabetes. 2016;65(4):902–12. doi:10.2337/db15-1094.
Walford GA, Davis J, Warner AS, Ackerman RJ, Billings LK, Chamarthi B, et al. Branched chain and aromatic amino acids change acutely following two medical therapies for type 2 diabetes mellitus. Metabolism. 2013;62(12):1772–8.
Laferrere B, Reilly D, Arias S, Swerdlow N, Gorroochurn P, Bawa B, et al. Differential metabolic impact of gastric bypass surgery versus dietary intervention in obese diabetic subjects despite identical weight loss. Sci Transl Med. 2011;3(80):80re2. doi:10.1126/scitranslmed.3002043.
Lyssenko V, Jonsson A, Almgren P, Pulizzi N, Isomaa B, Tuomi T, et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med. 2008;359(21):2220–32.
Floyd Jr JC, Fajans SS, Conn JW, Knopf RF, Rull J. Stimulation of insulin secretion by amino acids. J Clin Invest. 1966;45(9):1487–502.
van Loon LJ, Saris WH, Verhagen H, Wagenmakers AJ. Plasma insulin responses after ingestion of different amino acid or protein mixtures with carbohydrate. Am J Clin Nutr. 2000;72(1):96–105.
Newgard CB, Matschinsky FM. Substrate control of insulin release. Comprehensive Physiology. 2011;125–151. doi:10.1002/cphy.cp070205.
Nilsson M, Holst JJ, Bjorck IM. Metabolic effects of amino acid mixtures and whey protein in healthy subjects: studies using glucose-equivalent drinks. Am J Clin Nutr. 2007;85(4):996–1004.
Patti ME, Brambilla E, Luzi L, Landaker EJ, Kahn CR. Bidirectional modulation of insulin action by amino acids. J Clin Invest. 1998;101:1519–29.
Lynch CJ, Adams SH. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat Rev Endocrinol. 2014;10(12):723–36.
Macotela Y, Emanuelli B, Bang AM, Espinoza DO, Boucher J, Beebe K, et al. Dietary leucine—an environmental modifier of insulin resistance acting on multiple levels of metabolism. PLoS One. 2011;6(6):e21187.
She P, Reid TM, Bronson SK, Vary TC, Hajnal A, Lynch CJ, et al. Disruption of BCATm in mice leads to increased energy expenditure associated with the activation of a futile protein turnover cycle. Cell Metab. 2007;6(3):181–94.
Muoio DM, Newgard CB. Mechanisms of disease: molecular and metabolic mechanisms of insulin resistance and beta-cell failure in type 2 diabetes. Nat Rev Mol Cell Biol. 2008;9(3):193–205.
Morino K, Petersen KF, Shulman GI. Molecular mechanisms of insulin resistance in humans and their potential links with mitochondrial dysfunction. Diabetes. 2006;55 Suppl 2:S9–S15.
Boden G, Shulman GI. Free fatty acids in obesity and type 2 diabetes: defining their role in the development of insulin resistance and beta-cell dysfunction. Eur J Clin Invest. 2002;32 Suppl 3:14–23.
Lowell BB, Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science. 2005;307(5708):384–7.
Samuel VT, Petersen KF, Shulman GI. Lipid-induced insulin resistance: unravelling the mechanism. Lancet. 2010;375(9733):2267–77.
Dube JJ, Amati F, Toledo FG, Stefanovic-Racic M, Rossi A, Coen P, et al. Effects of weight loss and exercise on insulin resistance, and intramyocellular triacylglycerol, diacylglycerol and ceramide. Diabetologia. 2011;54(5):1147–56.
Itani SI, Ruderman NB, Schmieder F, Boden G. Lipid-induced insulin resistance in human muscle is associated with changes in diacylglycerol, protein kinase C, and IkappaB-alpha. Diabetes. 2002;51(7):2005–11.
Adams JM, Pratipanawatr T, Berria R, Wang E, DeFronzo RA, Sullards MC, et al. Ceramide content is increased in skeletal muscle from obese insulin-resistant humans. Diabetes. 2004;53(1):25–31.
Oh DY, Talukdar S, Bae EJ, Imamura T, Morinaga H, Fan W, et al. GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell. 2010;142(5):687–98.
Virtanen JK, Mursu J, Voutilainen S, Uusitupa M, Tuomainen TP. Serum omega-3 polyunsaturated fatty acids and risk of incident type 2 diabetes in men: the Kuopio Ischemic Heart Disease Risk Factor study. Diabetes Care. 2014;37(1):189–96.
Cao H, Gerhold K, Mayers JR, Wiest MM, Watkins SM, Hotamisligil GS. Identification of a lipokine, a lipid hormone linking adipose tissue to systemic metabolism. Cell. 2008;134(6):933–44.
Rhee EP, Cheng S, Larson MG, Walford GA, Lewis GD, McCabe E, et al. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J Clin Invest. 2011;121(4):1402–11. This article shows a relationship between lipid acyl chain content and diabetes risk (lower carbon number and double bond content was linked to increased risk), suggesting lipid profiles may identify high-risk individuals.
Riserus U, Willett WC, Hu FB. Dietary fats and prevention of type 2 diabetes. Prog Lipid Res. 2009;48(1):44–51.
Yore MM, Syed I, Moraes-Vieira PM, Zhang T, Herman MA, Homan EA, et al. Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell. 2014;159(2):318–32.
Illig T, Gieger C, Zhai G, Romisch-Margl W, Wang-Sattler R, Prehn C, et al. A genome-wide perspective of genetic variation in human metabolism. Nat Genet. 2010;42(2):137–41. The authors performed a GWAS of 163 metabolic traits in one large human cohort (with replication in a second one) and identified loci explaining 5.6–36.3 % of variance in metabolite concentrations.
Xie W, Wood AR, Lyssenko V, Weedon MN, Knowles JW, Alkayyali S, et al. Genetic variants associated with glycine metabolism and their role in insulin sensitivity and type 2 diabetes. Diabetes. 2013;62(6):2141–50.
Tannock GW. The normal microflora: new concepts in health promotion. Microbiol Sci. 1988;5(1):4–8.
Menni C, Zhai G, MacGregor A, Prehn C, Romisch-Margl W, Suhre K, et al. Targeted metabolomics profiles are strongly correlated with nutritional patterns in women. Metabolomics. 2013;9(2):506–14.
Roberts LD, Bostrom P, O’Sullivan JF, Schinzel RT, Lewis GD, Dejam A, et al. Beta-aminoisobutyric acid induces browning of white fat and hepatic beta-oxidation and is inversely correlated with cardiometabolic risk factors. Cell Metab. 2014;19(1):96–108.
Lawton KA, Berger A, Mitchell M, Milgram KE, Evans AM, Guo L, et al. Analysis of the adult human plasma metabolome. Pharmacogenomics. 2008;9(4):383–97.
Nicholson JK, Wilson ID. Opinion: understanding ‘global’ systems biology: metabonomics and the continuum of metabolism. Nat Rev Drug Discov. 2003;2(8):668–76.
Nicholson JK, Holmes E, Lindon JC, Wilson ID. The challenges of modeling mammalian biocomplexity. Nat Biotechnol. 2004;22(10):1268–74.
Wu H, Tremaroli V, Backhed F. Linking microbiota to human diseases: a systems biology perspective. Trends Endocrinol Metab. 2015;26(12):758–70.
Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–4.
Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102(31):11070–5.
Remely M, Dworzak S, Hippe B, Zwielehner J, Aumüller E, Brath H, et al. Abundance and diversity of microbiota in type 2 diabetes and obesity. Diabetes and Metabolism. 2013;4:3.
Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, Pedersen BK, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One. 2010;5(2):e9085.
Wu X, Ma C, Han L, Nawaz M, Gao F, Zhang X, et al. Molecular characterisation of the faecal microbiota in patients with type II diabetes. Curr Microbiol. 2010;61(1):69–78.
Zhang H, DiBaise JK, Zuccolo A, Kundrna D, Braidotti M, Yu Y, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci U S A. 2009;106(7):2365--70. doi: 10.1073/pnas.0812600106.
Holmes E, Li JV, Athanasiou T, Ashrafian H, Nicholson JK. Understanding the role of gut microbiome-host metabolic signal disruption in health and disease. Trends Microbiol. 2011;19(7):349–59.
Patti ME, Houten SM, Bianco AC, Bernier R, Larsen PR, Holst JJ, et al. Serum bile acids are higher in humans with prior gastric bypass: potential contribution to improved glucose and lipid metabolism. Obesity (Silver Spring). 2009;17(9):1671–7. doi:10.1038/oby.2009.102.
Mutch DM, Fuhrmann JC, Rein D, Wiemer JC, Bouillot JL, Poitou C, et al. Metabolite profiling identifies candidate markers reflecting the clinical adaptations associated with Roux-en-Y gastric bypass surgery. PLoS One. 2009;4(11):e7905.
Dumas ME, Barton RH, Toye A, Cloarec O, Blancher C, Rothwell A, et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci U SA. 2006;103(33):12511–6.
Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–31.
Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3(4):213–23.
Tremaroli V, Karlsson F, Werling M, Stahlman M, Kovatcheva-Datchary P, Olbers T, et al. Roux-en-Y gastric bypass and vertical banded gastroplasty induce long-term changes on the human gut microbiome contributing to fat mass regulation. Cell Metab. 2015;22(2):228–38.
Vrieze A, Van Nood E, Holleman F, Salojarvi J, Kootte RS, Bartelsman JF, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143(4):913–6. e7.
Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472(7341):57–63.
Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368(17):1575–84.
Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19(5):576–85.
Organ CL, Otsuka H, Bhushan S, Wang Z, Bradley J, Trivedi R, et al. Choline diet and its gut microbe-derived metabolite, trimethylamine N-oxide, exacerbate pressure overload-induced heart failure. Circ Heart Fail. 2016;9(1):e002314.
Sas KM, Karnovsky A, Michailidis G, Pennathur S. Metabolomics and diabetes: analytical and computational approaches. Diabetes. 2015;64(3):718–32.
Ferrara CT, Wang P, Neto EC, Stevens RD, Bain JR, Wenner BR, et al. Genetic networks of liver metabolism revealed by integration of metabolic and transcriptional profiling. PLoS Genet. 2008;4(3):e1000034.
Jerby L, Shlomi T, Ruppin E. Computational reconstruction of tissue-specific metabolic models: application to human liver metabolism. Mol Syst Biol. 2010;6:401.
Gille C, Bolling C, Hoppe A, Bulik S, Hoffmann S, Hubner K, et al. HepatoNet1: a comprehensive metabolic reconstruction of the human hepatocyte for the analysis of liver physiology. Mol Syst Biol. 2010;6:411.
Bordbar A, Feist AM, Usaite-Black R, Woodcock J, Palsson BO, Famili I. A multi-tissue type genome-scale metabolic network for analysis of whole-body systems physiology. BMC Syst Biol. 2011;5:180.
Nogiec C, Burkart A, Dreyfuss JM, Lerin C, Kasif S, Patti ME. Metabolic modeling of muscle metabolism identifies key reactions linked to insulin resistance phenotypes. Mol Metab. 2015;4(3):151–63.
Shlomi T, Cabili MN, Ruppin E. Predicting metabolic biomarkers of human inborn errors of metabolism. Mol SystBiol. 2009;5:263.
Kuc S, Koster MP, Pennings JL, Hankemeier T, Berger R, Harms AC, et al. Metabolomics profiling for identification of novel potential markers in early prediction of preeclampsia. PLoS One. 2014;9(5):e98540.
Maitre L, Fthenou E, Athersuch T, Coen M, Toledano MB, Holmes E, et al. Urinary metabolic profiles in early pregnancy are associated with preterm birth and fetal growth restriction in the Rhea mother-child cohort study. BMC Med. 2014;12:110.
de Seymour JV, Conlon CA, Sulek K, Villas Boas SG, McCowan LM, Kenny LC, et al. Early pregnancy metabolite profiling discovers a potential biomarker for the subsequent development of gestational diabetes mellitus. Acta Diabetol. 2014;51(5):887–90.
Ciborowski M, Zbucka-Kretowska M, Bomba-Opon D, Wielgos M, Brawura-Biskupski-Samaha R, Pierzynski P, et al. Potential first trimester metabolomic biomarkers of abnormal birth weight in healthy pregnancies. Prenat Diagn. 2014;34(9):870–7.
Heazell AE, Bernatavicius G, Warrander L, Brown MC, Dunn WB. A metabolomic approach identifies differences in maternal serum in third trimester pregnancies that end in poor perinatal outcome. Reprod Sci. 2012;19(8):863–75.
Bahado-Singh RO, Akolekar R, Mandal R, Dong E, Xia J, Kruger M, et al. First-trimester metabolomic detection of late-onset preeclampsia. Am J Obstet Gynecol. 2013;208(1):58–e1-7.
Horgan RP, Broadhurst DI, Walsh SK, Dunn WB, Brown M, Roberts CT, et al. Metabolic profiling uncovers a phenotypic signature of small for gestational age in early pregnancy. J Proteome Res. 2011;10(8):3660–73.
Huynh J, Xiong G, Bentley-Lewis R. A systematic review of metabolite profiling in gestational diabetes mellitus. Diabetologia. 2014;57(12):2453–64.
Metzger BE, Phelps RL, Freinkel N, Navickas IA. Effects of gestational diabetes on diurnal profiles of plasma glucose, lipids, and individual amino acids. Diabetes Care. 1980;3(3):402–9.
Scholtens DM, Muehlbauer MJ, Daya NR, Stevens RD, Dyer AR, Lowe LP, et al. Metabolomics reveals broad-scale metabolic perturbations in hyperglycemic mothers during pregnancy. Diabetes Care. 2014;37(1):158–66.
Pappa KI, Vlachos G, Theodora M, Roubelaki M, Angelidou K, Antsaklis A. Intermediate metabolism in association with the amino acid profile during the third trimester of normal pregnancy and diet-controlled gestational diabetes. Am J Obstet Gynecol. 2007;196(1):65–e1-5.
Diaz SO, Pinto J, Graca G, Duarte IF, Barros AS, Galhano E, et al. Metabolic biomarkers of prenatal disorders: an exploratory NMR metabonomics study of second trimester maternal urine and blood plasma. J Proteome Res. 2011;10(8):3732–42.
Dudzik D, Zorawski M, Skotnicki M, Zarzycki W, Kozlowska G, Bibik-Malinowska K, et al. Metabolic fingerprint of gestational diabetes mellitus. J Proteomics. 2014;103:57–71.
Pinto J, Almeida LM, Martins AS, Duarte D, Barros AS, Galhano E, et al. Prediction of gestational diabetes through NMR metabolomics of maternal blood. J Proteome Res. 2015;14(6):2696–706.
Cetin I, de Santis MS, Taricco E, Radaelli T, Teng C, Ronzoni S, et al. Maternal and fetal amino acid concentrations in normal pregnancies and in pregnancies with gestational diabetes mellitus. Am J Obstet Gynecol. 2005;192(2):610–7.
Lehmann R, Friedrich T, Krebiehl G, Sonntag D, Haring HU, Fritsche A, et al. Metabolic profiles during an oral glucose tolerance test in pregnant women with and without gestational diabetes. Exp Clin Endocrinol Diabetes. 2015;123(7):483–38.
Prentice KJ, Luu L, Allister EM, Liu Y, Jun LS, Sloop KW, et al. The furan fatty acid metabolite CMPF is elevated in diabetes and induces beta cell dysfunction. Cell Metab. 2014;19(4):653–66.
Dani C, Bresci C, Berti E, Ottanelli S, Mello G, Mecacci F, et al. Metabolomic profile of term infants of gestational diabetic mothers. J Matern Fetal Neonatal Med. 2014;27(6):537–42.
Gluckman PD, Hanson MA, Cooper C, Thornburg KL. Effect of in utero and early-life conditions on adult health and disease. N Engl J Med. 2008;359(1):61–73.
Wang G, Divall S, Radovick S, Paige D, Ning Y, Chen Z, et al. Preterm birth and random plasma insulin levels at birth and in early childhood. JAMA. 2014;311(6):587–96.
Dessi A, Atzori L, Noto A, Visser GH, Gazzolo D, Zanardo V, et al. Metabolomics in newborns with intrauterine growth retardation (IUGR): urine reveals markers of metabolic syndrome. J Matern Fetal Neonatal Med. 2011;24 Suppl 2:35–9.
Ivorra C, Garcia-Vicent C, Chaves FJ, Monleon D, Morales JM, Lurbe E. Metabolomic profiling in blood from umbilical cords of low birth weight newborns. J Transl Med. 2012;10:142. doi:10.1186/1479-5876-10-142.
Alexandre-Gouabau MC, Courant F, Moyon T, Kuster A, Le GG, Tea I, et al. Maternal and cord blood LC-HRMS metabolomics reveal alterations in energy and polyamine metabolism, and oxidative stress in very-low birth weight infants. J Proteome Res. 2013;12(6):2764–78.
Leunissen RW, Kerkhof GF, Stijnen T, Hokken-Koelega A. Timing and tempo of first-year rapid growth in relation to cardiovascular and metabolic risk profile in early adulthood. JAMA. 2009;301(21):2234–42.
Barker DJ, Osmond C, Forsen TJ, Kajantie E, Eriksson JG. Trajectories of growth among children who have coronary events as adults. New England Journal of Medicine. 2005;353(17):1802–9.
Isganaitis E, Rifas-Shiman SL, Oken E, Dreyfuss JM, Gall W, Gillman MW, et al. Associations of cord blood metabolites with early childhood obesity risk. Int J Obes (Lond). 2015;39(7):1041–8.
Rzehak P, Hellmuth C, Uhl O, Kirchberg FF, Peissner W, Harder U, et al. Rapid growth and childhood obesity are strongly associated with lysoPC(14:0). Ann Nutr Metab. 2014;64(3–4):294–303.
Stevens A, Bonshek C, Whatmore A, Butcher I, Hanson D, De Leonibus C, et al. Insights into the pathophysiology of catch-up compared with non-catch-up growth in children born small for gestational age: an integrated analysis of metabolic and transcriptomic data. Pharmacogenomics J. 2014;14(4):376–84.
Perng W, Gillman MW, Fleisch AF, Michalek RD, Watkins SM, Isganaitis E, et al. Metabolomic profiles and childhood obesity. Obesity (Silver Spring). 2014;22(12):2570–8.
Mihalik SJ, Michaliszyn SF, de las Heras J, Bacha F, Chace DH, et al. Metabolomic profiling of fatty acid and amino acid metabolism in youth with obesity and type 2 diabetes: evidence for enhanced mitochondrial oxidation. Diabetes Care. 2012;35(3):605–11.
Butte NF, Liu Y, Zakeri IF, Mohney RP, Mehta N, Voruganti VS, et al. Global metabolomic profiling targeting childhood obesity in the Hispanic population. Am J Clin Nutr. 2015;102(2):256–67.
McCormack SE, Shaham O, McCarthy MA, Deik AA, Wang TJ, Gerszten RE, et al. Circulating branched-chain amino acid concentrations are associated with obesity and future insulin resistance in children and adolescents. Pediatr Obes. 2013;8(1):52–61.
Wahl S, Yu Z, Kleber M, Singmann P, Holzapfel C, He Y, et al. Childhood obesity is associated with changes in the serum metabolite profile. Obes Facts. 2012;5(5):660–70.
Michaliszyn SF, Sjaarda LA, Mihalik SJ, Lee S, Bacha F, Chace DH, et al. Metabolomic profiling of amino acids and beta-cell function relative to insulin sensitivity in youth. J Clin Endocrinol Metab. 2012;97(11):E2119–E24.
Acknowledgments
AG-F gratefully acknowledges support from the American Diabetes Association (mentored research fellowship to MEP). AMB acknowledges research support from the NIH (T32 DK 007260) and the Harold Whitworth Pierce Charitable Trust Postdoctoral Fellowship. EI acknowledges grant support from K99R00 HD064793 and Boston Nutrition and Obesity Research Center. MEP acknowledges research support from the NIH, American Diabetes Association, Bristol-Myers Squibb, Janssen, Nuclea, Medimmune, and Joslin DRC grant P30 DK036836.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Alba Gonzalez-Franquesa, Alison M. Burkart, Elvira Isganaitis, and Mary-Elizabeth Patti declare no conflict of interest related to this manuscript.
Human and Animal Rights and Informed Consent
This article does not report any new studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Pathogenesis of Type 2 Diabetes and Insulin Resistance
Rights and permissions
About this article
Cite this article
Gonzalez-Franquesa, A., Burkart, A.M., Isganaitis, E. et al. What Have Metabolomics Approaches Taught Us About Type 2 Diabetes?. Curr Diab Rep 16, 74 (2016). https://doi.org/10.1007/s11892-016-0763-1
Published:
DOI: https://doi.org/10.1007/s11892-016-0763-1