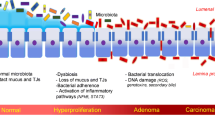
Abstract
Purpose of Review
Studies have identified differences between the gut microbiota of colorectal cancer (CRC) patients versus healthy individuals. In this review, we assess the scientific literature to determine if gut microbes should be considered causal, co-varying, or a necessary but not sufficient agent in CRC development.
Recent Findings
Oral bacteria may influence CRC susceptibility. Colonic biofilms in both sporadic and hereditary CRC suggest these bacteria are present in early neoplasia. Pathogenic drivers and opportunistic passenger bacteria may underlie direct effect of the gut microbiota on carcinogenesis.
Summary
Members of multiple bacterial taxa have been implicated in CRC tumorigenesis and progression, with distinct mechanisms of action described for each. Individual bacterial organisms found in the colon are likely not enough to explain CRC development and progression. The entire colonic environment, including genetic factors, local tissue inflammatory state as well as dietary components may influence the way epithelial cells respond to the presence of certain bacteria. Longitudinal, human intervention studies are needed to completely clarify complex interactions in the colonic environment and specific causative pathways between the microbiota and CRC.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Dolatkhah R, Somi MH, Shabanloei R, Farassati F, Fakhari A, Dastgiri S. Main risk factors association with proto-oncogene mutations in colorectal cancer. Asian Pac J Cancer Prev. 2018;19(8):2183–90. https://doi.org/10.22034/APJCP.2018.19.8.2183.
Yang J, Yu J. The association of diet, gut microbiota and colorectal cancer: what we eat may imply what we get. Protein Cell. 2018;9(5):474–87. https://doi.org/10.1007/s13238-018-0543-6.
Carr PR, Weigl K, Jansen L, Walter V, Erben V, Chang-Claude J, et al. Healthy lifestyle factors associated with lower risk of colorectal cancer irrespective of genetic risk. Gastroenterology. 2018;155(6):1805–15 e5. https://doi.org/10.1053/j.gastro.2018.08.044.
Son JS, Khair S, Pettet DW, Ouyang NT, Tian XY, Zhang YH, et al. Altered interactions between the gut microbiome and colonic mucosa precede polyposis in APC(min/+) mice. PLoS One. 2015;10(6):e0127985. https://doi.org/10.1371/journal.pone.0127985.
Bhatt AP, Redinbo MR, Bultman SJ. The role of the microbiome in cancer development and therapy. CA Cancer J Clin. 2017;67(4):326–44. https://doi.org/10.3322/caac.21398.
Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One. 2012;7(6):e39743. https://doi.org/10.1371/journal.pone.0039743.
Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P, et al. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One. 2011;6(1):e16393. https://doi.org/10.1371/journal.pone.0016393.
Vogtmann E, Hua X, Zeller G, Sunagawa S, Voigt AY, Hercog R, et al. Colorectal cancer and the human gut microbiome: reproducibility with whole-genome shotgun sequencing. PLoS One. 2016;11(5):e0155362. https://doi.org/10.1371/journal.pone.0155362.
Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2017;66(1):70–8. https://doi.org/10.1136/gutjnl-2015-309800.
Sze MA, Schloss PD. Leveraging existing 16S rRNA gene surveys to identify reproducible biomarkers in individuals with colorectal tumors (vol 9, e00630–18, 2018). Mbio. 2018;9(5):e02076–18. https://doi.org/10.1128/mBio.02076-18.
Shah MS, DeSantis TZ, Weinmaier T, McMurdie PJ, Cope JL, Altrichter A, et al. Leveraging sequence-based faecal microbial community survey data to identify a composite biomarker for colorectal cancer. Gut. 2018;67(5):882–91. https://doi.org/10.1136/gutjnl-2016-313189.
Liang QY, Chiu J, Chen YX, Huang YQ, Higashimori A, Fang JY, et al. Fecal Bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clin Cancer Res. 2017;23(8):2061–70. https://doi.org/10.1158/1078-0432.Ccr-16-1599.
Dai ZW, Coker OO, Nakatsu G, Wu WK, Zhao LY, Chen ZG, et al. Multi-cohort analysis of colorectal cancer metagenome identified altered Bacteria across populations and universal bacterial markers. Gastroenterology. 2018;154(6):S1047–S8.
Zackular JP, Rogers MA, Ruffin MT, Schloss PD. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev Res (Phila). 2014;7(11):1112–21. https://doi.org/10.1158/1940-6207.CAPR-14-0129.
Flemer B, Warren RD, Barrett MP, Cisek K, Das A, Jeffery IB, et al. The oral microbiota in colorectal cancer is distinctive and predictive. Gut. 2018;67(8):1454–63. https://doi.org/10.1136/gutjnl-2017-314814.
• Kwong TNY, Wang X, Nakatsu G, Chow TC, Tipoe T, Dai RZW, et al. Association between bacteremia from specific microbes and subsequent diagnosis of colorectal cancer. Gastroenterology. 2018;155(2):383–90 e8. https://doi.org/10.1053/j.gastro.2018.04.028 A large-scale cohort study showing an association between bacteremia with CRC-associated bacteria and subsequent CRC diagnosis.
Gao R, Kong C, Li H, Huang L, Qu X, Qin N, et al. Dysbiosis signature of mycobiota in colon polyp and colorectal cancer. Eur J Clin Microbiol Infect Dis. 2017;36(12):2457–68. https://doi.org/10.1007/s10096-017-3085-6.
• Hannigan GD, Duhaime MB, Ruffin MT, Koumpouras CC, Schloss PD. Diagnostic potential and interactive dynamics of the colorectal cancer virome. MBio. 2018;9(6). https://doi.org/10.1128/mBio.02248-18 Presents a novel model for predicting CRC based on bacteriophages present in the gut and presents evidence that bacteriophages may impact CRC progression by modulating gut bacteria.
Puppa MJ, White JP, Sato S, Cairns M, Baynes JW, Carson JA. Gut barrier dysfunction in the ApcMin/+ mouse model of colon cancer cachexia. BBA-Mol Basis Dis. 2011;1812(12):1601–6. https://doi.org/10.1016/j.bbadis.2011.08.010.
Bornholdt J, Friis S, Godiksen S, Poulsen SS, Santoni-Rugiu E, Bisgaard HC, et al. The level of claudin-7 is reduced as an early event in colorectal carcinogenesis. BMC Cancer. 2011;11:65. https://doi.org/10.1186/1471-2407-11-65.
Luissint AC, Parkos CA, Nusrat A. Inflammation and the intestinal barrier: leukocyte-epithelial cell interactions, cell junction remodeling, and mucosal repair. Gastroenterology. 2016;151(4):616–32. https://doi.org/10.1053/j.gastro.2016.07.008.
Lechuga S, Ivanov AI. Disruption of the epithelial barrier during intestinal inflammation: quest for new molecules and mechanisms. BBA-Mol Cell Res. 2017;1864(7):1183–94. https://doi.org/10.1016/j.bbamcr.2017.03.007.
Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. 2018;359(6375):592–7. https://doi.org/10.1126/science.aah3648.
Singh R, Chandrashekharappa S, Bodduluri SR, Baby BV, Hegde B, Kotla NG, et al. Enhancement of the gut barrier integrity by a microbial metabolite through the Nrf2 pathway. Nat Commun. 2019;10:89. https://doi.org/10.1038/s41467-018-07859-7.
Bhutiani N, Li QS, Anderson CD, Gallagher HC, De Jesus M, Singh R, et al. Enhanced gut barrier integrity sensitizes colon cancer to immune therapy. Oncoimmunology. 2018;7(11). https://doi.org/10.1080/2162402x.2018.1498438.
Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun. 2015;6:6528. https://doi.org/10.1038/ncomms7528.
Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10(8):575–82. https://doi.org/10.1038/nrmicro2819.
Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15(9):1016–22. https://doi.org/10.1038/nm.2015.
•• Purcell RV, Pearson J, Aitchison A, Dixon L, Frizelle FA, Keenan JI. Colonization with enterotoxigenic Bacteroides fragilis is associated with early-stage colorectal neoplasia. PLoS One. 2017;12(2):e0171602. https://doi.org/10.1371/journal.pone.0171602 Documents the detection of the proposed CRC-associated driver pathogen, ETBF, at the earliest stages of neoplasia, suggesting a role for ETBF in initiation of CRC.
Amitay EL, Krilaviciute A, Brenner H. Systematic review: gut microbiota in fecal samples and detection of colorectal neoplasms. Gut Microbes. 2018;9(4):293–307. https://doi.org/10.1080/19490976.2018.1445957.
Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170(3):548–63 e16. https://doi.org/10.1016/j.cell.2017.07.008.
Viljoen KS, Dakshinamurthy A, Goldberg P, Blackburn JM. Quantitative profiling of colorectal cancer-associated bacteria reveals associations between fusobacterium spp., enterotoxigenic Bacteroides fragilis (ETBF) and clinicopathological features of colorectal cancer. PLoS One. 2015;10(3):e0119462. https://doi.org/10.1371/journal.pone.0119462.
Abed J, Emgard JE, Zamir G, Faroja M, Almogy G, Grenov A, et al. Fap2 mediates fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe. 2016;20(2):215–25. https://doi.org/10.1016/j.chom.2016.07.006.
Shang FM, Liu HL. Fusobacterium nucleatum and colorectal cancer: a review. World J Gastrointest Oncol. 2018;10(3):71–81. https://doi.org/10.4251/wjgo.v10.i3.71.
Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-kappaB, and up-regulating expression of microRNA-21. Gastroenterology. 2017;152(4):851–66 e24. https://doi.org/10.1053/j.gastro.2016.11.018.
•• Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358(6369):1443–8. https://doi.org/10.1126/science.aal5240 Demonstrates that viable, CRC-associated bacteria travel with primary CRC cells to distant organs through metastasis. They further demonstrated reduction of tumor growth by antibiotic treatment.
Kosumi K, Hamada T, Koh H, Borowsky J, Bullman S, Twombly TS, et al. The amount of Bifidobacterium genus in colorectal carcinoma tissue in relation to tumor characteristics and clinical outcome. Am J Pathol. 2018;188(12):2839–52. https://doi.org/10.1016/j.ajpath.2018.08.015.
Sivaprakasam S, Gurav A, Paschall AV, Coe GL, Chaudhary K, Cai Y, et al. An essential role of Ffar2 (Gpr43) in dietary fibre-mediated promotion of healthy composition of gut microbiota and suppression of intestinal carcinogenesis. Oncogene. 2016;5(6):e238. https://doi.org/10.1038/oncsis.2016.38.
Brim H, Yooseph S, Lee E, Sherif ZA, Abbas M, Laiyemo AO, et al. A microbiomic analysis in African Americans with colonic lesions reveals Streptococcus sp.VT162 as a marker of neoplastic transformation. Genes. 2017;8(11):314. https://doi.org/10.3390/genes8110314.
Bultman SJ. Interplay between diet, gut microbiota, epigenetic events, and colorectal cancer. Mol Nutr Food Res. 2017;61(1):1500902. https://doi.org/10.1002/mnfr.201500902.
Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. Diet-induced extinctions in the gut microbiota compound over generations. Nature. 2016;529(7585):212–U08. https://doi.org/10.1038/nature16504.
Qamar TR, Syed F, Nasir M, Rehman H, Zahid MN, Liu RH, et al. Novel combination of prebiotics galacto-oligosaccharides and inulin-inhibited aberrant crypt foci formation and biomarkers of colon cancer in wistar rats. Nutrients. 2016;8(8). https://doi.org/10.3390/nu8080465.
Cao HL, Xu MQ, Dong WX, Deng BR, Wang SN, Zhang YJ, et al. Secondary bile acid-induced dysbiosis promotes intestinal carcinogenesis. Int J Cancer. 2017;140(11):2545–56. https://doi.org/10.1002/ijc.30643.
O'Keefe SJ, Li JV, Lahti L, Ou J, Carbonero F, Mohammed K, et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun. 2015;6:6342. https://doi.org/10.1038/ncomms7342.
Sheflin AM, Borresen EC, Kirkwood JS, Boot CM, Whitney AK, Lu S, et al. Dietary supplementation with rice bran or navy bean alters gut bacterial metabolism in colorectal cancer survivors. Mol Nutr Food Res. 2017;61(1). https://doi.org/10.1002/mnfr.201500905.
Nurdin SU, Le Leu RK, Young GP, Stangoulis JCR, Christophersen CT, Abbott CA. Analysis of the anti-cancer effects of cincau extract (Premna oblongifolia merr) and other types of non-digestible fibre using faecal fermentation supernatants and Caco-2 cells as a model of the human colon. Nutrients. 2017;9(4):355. https://doi.org/10.3390/nu9040355.
van der Beek CM, Dejong CHC, Troost FJ, Masclee AM, Lenaerts K. Role of short-chain fatty acids in colonic inflammation, carcinogenesis, and mucosal protection and healing. Nutr Rev. 2017;75(4):286–305. https://doi.org/10.1093/nutrit/nuw067.
Han RR, Sun QQ, Wu JB, Zheng PY, Zhao GQ. Sodium butyrate upregulates miR-203 expression to exert anti-proliferation effect on colorectal cancer cells. Cell Physiol Biochem. 2016;39(5):1919–29. https://doi.org/10.1159/000447889.
Bishehsari F, Engen PA, Preite NZ, Tuncil YE, Naqib A, Shaikh M, et al. Dietary fiber treatment corrects the composition of gut microbiota, promotes SCFA production, and suppresses colon carcinogenesis. Genes. 2018;9(2):102. https://doi.org/10.3390/genes9020102.
Nowak A, Slizewska K, Otlewska A. Antigenotoxic activity of lactic acid bacteria, prebiotics, and products of their fermentation against selected mutagens. Regul Toxicol Pharmacol. 2015;73(3):938–46. https://doi.org/10.1016/j.yrtph.2015.09.021.
Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165(6):1332–45. https://doi.org/10.1016/j.cell.2016.05.041.
Kang HR, Choi HG, Jeon CK, Lim SJ, Kim SH. Butyrate-mediated acquisition of chemoresistance by human colon cancer cells. Oncol Rep. 2016;36(2):1119–26. https://doi.org/10.3892/or.2016.4838.
Bardhan K, Paschall AV, Yang D, Chen MR, Simon PS, Bhutia YD, et al. IFNgamma induces DNA methylation-silenced GPR109A expression via pSTAT1/p300 and H3K18 acetylation in colon cancer. Cancer Immunol Res. 2015;3(7):795–805. https://doi.org/10.1158/2326-6066.CIR-14-0164.
Sivaprakasam S, Prasad PD, Singh N. Benefits of short-chain fatty acids and their receptors in inflammation and carcinogenesis. Pharmacol Ther. 2016;164:144–51. https://doi.org/10.1016/j.pharmthera.2016.04.007.
Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350(6264):1084–9. https://doi.org/10.1126/science.aac4255.
Man SM, Zhu Q, Zhu L, Liu Z, Karki R, Malik A, et al. Critical role for the DNA sensor AIM2 in stem cell proliferation and cancer. Cell. 2015;162(1):45–58. https://doi.org/10.1016/j.cell.2015.06.001.
Killeen SD, Wang JH, Andrews EJ, Redmond HP. Bacterial endotoxin enhances colorectal cancer cell adhesion and invasion through TLR-4 and NF-kappa B-dependent activation of the urokinase plasminogen activator system. Br J Cancer. 2009;100(10):1589–602. https://doi.org/10.1038/sj.bjc.6604942.
Hsu RY, Chan CH, Spicer JD, Rousseau MC, Giannias B, Rousseau S, et al. LPS-induced TLR4 signaling in human colorectal cancer cells increases beta1 integrin-mediated cell adhesion and liver metastasis. Cancer Res. 2011;71(5):1989–98. https://doi.org/10.1158/0008-5472.CAN-10-2833.
Zhu G, Huang Q, Huang Y, Zheng W, Hua J, Yang S, et al. Lipopolysaccharide increases the release of VEGF-C that enhances cell motility and promotes lymphangiogenesis and lymphatic metastasis through the TLR4- NF-kappaB/JNK pathways in colorectal cancer. Oncotarget. 2016;7(45):73711–24. https://doi.org/10.18632/oncotarget.12449.
Beaugerie L, Itzkowitz SH. Cancers complicating inflammatory bowel disease reply. N Engl J Med. 2015;373(2):195.
Brennan CA, Garrett WS. Gut microbiota, inflammation, and colorectal cancer. Annu Rev Microbiol. 2016;70:395–411. https://doi.org/10.1146/annurev-micro-102215-095513.
Kostic AD, Chun EY, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207–15. https://doi.org/10.1016/j.chom.2013.07.007.
•• Aymeric L, Donnadieu F, Mulet C, du Merle L, Nigro G, Saffarian A, et al. Colorectal cancer specific conditions promote Streptococcus gallolyticus gut colonization. Proc Natl Acad Sci U S A. 2018;115(2):E283–E91. https://doi.org/10.1073/pnas.1715112115 A description of intestinal colonization by an early CRC-associated microbe. The mechanism suggests a colonization following tumorigenesis (rather than preceding), and connects host genetics, bile-acid accumulation, and bacterial virulence factors, all previously associated with CRC.
Wong SH, Zhao LY, Zhang X, Nakatsu G, Han JQ, Xu WQ, et al. Gavage of fecal samples from patients with colorectal cancer promotes intestinal carcinogenesis in germ-free and conventional mice. Gastroenterology. 2017;153(6):1621–1633.e6. https://doi.org/10.1053/j.gastro.2017.08.022.
Barthold SW, Jonas AM. Morphogenesis of early 1, 2-dimethylhydrazine-induced lesions and latent period reduction of colon carcinogenesis in mice by a variant of Citrobacter freundii. Cancer Res. 1977;37(12):4352–60.
Newman JV, Kosaka T, Sheppard BJ, Fox JG, Schauer DB. Bacterial infection promotes colon tumorigenesis in Apc(min/+) mice. J Infect Dis. 2001;184(2):227–30. https://doi.org/10.1086/321998.
Chandrakesan P, Ahmed I, Anwar T, Wang Y, Sarkar S, Singh P, et al. Novel changes in NF-{kappa}B activity during progression and regression phases of hyperplasia: role of MEK, ERK, and p38. J Biol Chem. 2010;285(43):33485–98. https://doi.org/10.1074/jbc.M110.129353.
Ahmed I, Chandrakesan P, Tawfik O, Xia L, Anant S, Umar S. Critical roles of Notch and Wnt/beta-catenin pathways in the regulation of hyperplasia and/or colitis in response to bacterial infection. Infect Immun. 2012;80(9):3107–21. https://doi.org/10.1128/IAI.00236-12.
Becker S, Oelschlaeger TA, Wullaert A, Pasparakis M, Wehkamp J, Stange EF, et al. Bacteria regulate intestinal epithelial cell differentiation factors both in vitro and in vivo. PLoS One. 2013;8(2):e55620. https://doi.org/10.1371/journal.pone.0055620.
Priyamvada S, Anbazhagan A, Chatterjee I, Alrefai W, Dudeja P, Borthakur A. Gut bacterial metabolite propionate upregulates intestinal epithelial kruppel-like factor 4 expression via a PPAR-y-dependent mechanism. FASEB J. 2015;29(1_supplement):854–4.
Zackular JP, Baxter NT, Chen GY, Schloss PD. Manipulation of the gut microbiota reveals role in colon tumorigenesis. Msphere. 2016;1(1):e00001–15. https://doi.org/10.1128/mSphere.00001-15.
Yu ZD, Song G, Liu J, Wang JY, Zhang PY, Chen KS. Beneficial effects of extracellular polysaccharide from Rhizopus nigricans on the intestinal immunity of colorectal cancer mice. Int J Biol Macromol. 2018;115:718–26. https://doi.org/10.1016/j.ijbiomac.2018.04.128.
Belcheva A, Irrazabal T, Robertson SJ, Streutker C, Maughan H, Rubino S, et al. Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell. 2014;158(2):288–99. https://doi.org/10.1016/j.cell.2014.04.051.
Acknowledgments
The authors would like to thank Drs. Christina Hester and Ishfaq Ahmed for thoughtful discussion.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Kristina M. Bridges, K. Allen Greiner, and Shahid Umar declare they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Basic Science Foundations in Colorectal Cancer
Rights and permissions
About this article
Cite this article
Bridges, K.M., Greiner, K.A. & Umar, S. Deciphering the Colorectal Cancer Gut Microbiota: Association vs. Causality. Curr Colorectal Cancer Rep 15, 70–77 (2019). https://doi.org/10.1007/s11888-019-00431-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11888-019-00431-5