Abstract
Phytochemicals not only determine the taste and smell of plants and their products, they also play a crucial role in resistance against pests and pathogens. In previous work, we identified a form of resistance to thrips (Frankliniella occidentalis) expressed in leaves of pepper (Capsicum annuum). In the current study, we characterized leaves of an interspecific C. annuum × C. chinense F2 population for variation in their global phytochemical composition by an untargeted metabolomics approach. Quantitative trait locus (QTL) mapping resulted in metabolite QTLs (mQTLs) for 304 of the 674 metabolites detected. We compared the QTL mapping results to those obtained earlier on fruits in the same population. Very different QTL hotspots were found, suggesting that the metabolite composition of leaves and fruits is regulated independently. Six leaf mQTLs co-located with the major QTL for resistance to thrips, which we previously identified in the same F2 population. Four of them were significantly correlated to thrips resistance, including two diterpene glycosides and a flavonoid compound which may indicate a possible role of these metabolites in thrips resistance. If a causal role of some of these metabolites in resistance can be proven this will help in the identification of the causal gene(s) and it may provide leads for the identification of other sources of thrips resistance in Capsicum and in other species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Peppers (Capsicum spp.) are among the most cultivated vegetables in the world today (FAOSTAT 2016). They can be used as food product, fresh or as cooked vegetable, as herb or spice, or in processed form. Peppers are also used in pharmacology (Bosland et al. 2012), as ornamental plant (Stummel and Bosland 2007), and even in protective (pepper) sprays (Miller 2001).
The quality of pepper fruits is determined by characteristics such as color, shape, size, uniformity, shelf life, taste, and biochemical composition. Fruits of pepper contain high levels of nutritional compounds, including carotenoids (provitamin A), ascorbic acid (vitamin C), tocopherols (vitamin E), phenolic compounds, flavonoids, and capsaicinoids (Howard and Wildman 2007; Topuz and Ozdemir 2007; Wahyuni et al. 2013a, b). In contrast to pepper fruits, no comprehensive metabolite profiling has yet been published for pepper leaves.
Pepper cultivation is hampered by several insect pests and thrips are among the most damaging; they cause damage directly by feeding on leaves, fruits, or flowers and indirectly by transmission of viruses, especially Tomato Spotted Wilt Virus (TSVW) (Ullman et al. 2002). Resistance to thrips (Frankliniella occidentalis) has been identified in Capsicum (Fery and Schalk 1991; Maris et al. 2003; Maharijaya et al. 2011). In general, it may be based on antixenosis (non-preference) or antibiosis (factors that increase mortality or reduce the growth, development, and reproduction of insects). Resistance components may interfere with any stage in the life cycle of the insect. For pepper, it was shown that resistance factors affected (blocked) the transition of first instar larvae (L1) into the second instar (L2), as determined in detached leaf assays (Maharijaya et al. 2012). In an earlier study (Maharijaya et al. 2011), it was already shown that resistance as determined by tests with young, detached leaves correlated well with resistance as determined in greenhouse tests. Although adult thrips are often found in flowers, as they feed on the pollen produced (Trichilo and Leigh 1988), they deposit their eggs mainly in the young leaves (Kiers et al. 2000; Van Haperen et al. submitted). So, resistance in leaves may affect thrips population development as the young larvae first feed on the leaves. Maharijaya et al. (2015) identified a major quantitative trait locus (QTL) for thrips resistance in an F2 population derived from an interspecific Capsicum annuum × C. chinense cross. This QTL was mapped on chromosome 6 and was shown to affect larval development of the insect, especially the transition from larval stage L1 to stage L2 and the pre-pupa stage on leaves (Maharijaya et al. 2012, 2015).
In several host species, resistance to thrips was found to be correlated with the presence of secondary metabolites (Blauth et al. 1998; Abe et al. 2008; Leiss et al. 2009a, b; Maharijaya et al. 2012; Vosman et al. 2018). Often it is not known why this correlation is there. Some suggestions have been made. In tomato for instance, the resistance was suggested to be based on the stickiness of the leaves of resistant plants, which may be due to the presence of large amounts of acyl sugars (Vosman et al. 2018). Co-location of QTLs for metabolites (mQTLs) with QTLs for thrips resistance can provide additional evidence that a compound may be involved in the resistance mechanism.
Gas chromatography–mass spectrometry (GC–MS) and liquid chromatography–mass spectrometry (LC–MS) are currently the standard methods for metabolite profiling (Dunn et al. 2013; Vinaixa et al. 2016). The exploitation of GC–MS and LC–MS data in an untargeted metabolomics approach allows the detection of hundreds of metabolites simultaneously, without prior knowledge on their identity, which may then be linked to genetic or phenotypic variation in order to select and subsequently identify compounds of most interest (Tikunov et al. 2005; Dunn et al. 2013; Alonso et al. 2015). An untargeted metabolomics approach is therefore very suitable for the detection and identification of those metabolites correlating to a specific trait. Since the resistance we mapped earlier was of the antibiosis type (Maharijaya et al. 2012), we used untargeted profiling of non-volatile phytochemicals, rather than volatile compounds, because non-volatiles are likely to be involved in the antibiosis type of thrips resistance in pepper (Wink 1988; Smith 2005).
The same pepper F2 population in which we previously mapped thrips resistance QTL (Maharijaya et al. 2015) has been used for untargeted LC–MS-based profiling and QTL mapping of fruit metabolites (Wahyuni et al. 2014). In these pepper fruits, a total of 542 metabolites, including series of flavonoids, phenylpropanoids, capsinoids, fatty acid derivatives, and amino acid derivatives, were detected and the QTL mapping resulted in the identification of 279 metabolite QTLs (mQTLs), dispersed over all 12 chromosomes with two hotspots on chromosome 9 (Wahyuni et al. 2014). The aims of our current study were (1) to determine metabolite profiles in leaves of pepper, detectable by LC–MS, and to map these in the F2 population; (2) to compare the distribution of mQTLs found in leaves with those in fruits as detected by (Wahyuni et al. 2014); and (3) to study the co-location of leaf mQTLs with the main thrips (Frankliniella occidentalis) resistance QTL on chromosome 6 (Maharijaya et al. 2015) in order to identify metabolites that are potentially involved in thrips resistance in pepper.
Materials and methods
Plant material
An F2 population consisting of 196 plants was developed from a cross between C. annuum AC 1979 as female parent and C. chinense 4661 as male parent (Maharijaya et al. 2015). The two parents differ in laboratory and greenhouse tests for their resistance level against two thrips species, Frankliniella occidentalis and Thrips parvispinus (Maharijaya et al. 2011). The maternal parent, C. annuum AC 1979 was very resistant to thrips and suppressed the development of L1 larvae while the paternal parent, C. chinense 4661 was very susceptible to thrips and supported the development of larvae (Maharijaya et al. 2012). Both accessions were obtained from the Center of Genetic Resources, the Netherlands. The F2 population was grown together with two first-generation inbred lines obtained by self-pollination of the two parental plants and with cuttings of the F1 plant in a glasshouse at Wageningen University and Research Center, the Netherlands in a completely randomized experiment. Mapping of thrips resistance in the F2 population is described by Maharijaya et al. (2015), which contains all details on the thrips rearing and assays used; damage caused by thrips larvae and the survival of first and second instar larval stages were used as parameters of resistance in a no-choice assay. The plants were maintained in standard glasshouse cultivation for pepper at 25 °C, with a daily photoperiod of 16-h light: 8-h dark (LD 16:8). Pest insects were controlled biologically using the predator Amblyseius sp. (Koppert®), to prevent that other herbivores would affect the resistance level against thrips or the metabolite profile.
Extraction and LC–MS analysis of pepper leaf metabolites
Young, fully opened, undamaged leaves of 13-week-old plants were harvested for metabolite analysis during the resistance evaluation (Maharijaya et al. 2015); this leaf stage corresponded to that used for the resistance evaluation. The plants (and these leaves) had not been in contact with thrips and metabolites detected are therefore to be considered as constitutively produced. The leaves were ground in liquid nitrogen into a fine powder. Five hundred milligram of the powder was weighed in a reaction tube and 1.5 ml 99.9% methanol acidified with 0.125% formic acid was added. Extracts were sonicated for 15 min and centrifuged for 5 min at 1515 g. Next, the supernatant was filtered through 0.2 µm polytetrafluoroethylene filter. All the extracts were analyzed on a reversed phase liquid chromatograph coupled to a photodiode array detector and a high-resolution mass spectrometer (LC–PDA–QTOF–MS) system (waters), using negative electrospray ionization, as described by De Vos et al. (2007).
LC–MS data analysis and putative metabolite annotation
An untargeted approach was applied to process the raw LC–MS data. Raw data were first processed by the MetAlign software package (Lommen 2009) for baseline correction, mass peak picking, and ion-wise alignment. The MetAlign output (peak heights) was processed with MSClust to cluster all individual ions originating from the same metabolite into the so-called centrotypes (Tikunov et al. 2012). Since each centrotype putatively represents an individual metabolite, in the following sections, these centrotypes are referred to as metabolites. Metabolites were putatively identified by comparing the retention times and molecular mass of detected compounds with that of two metabolite online databases, i.e., the Dictionary of Natural Products (ChemnetBase 2013) and KNApSAck (Nakamura et al. 2008), and with results on pepper fruit metabolite analyses as published by Marin et al. (2004) and Wahyuni et al. (2011, 2014).
QTL mapping of leaf and fruit metabolites
QTL mapping was performed in the F2 population described by Maharijaya et al. (2015), for which a linkage map composed of SSR, SNP, and AFLP markers was available. This linkage map consisted of 22 linkage groups, 16 of which were assigned to physical chromosomes. The total length of the linkage map was 1630 cM. We mapped mQTLs for the leaf metabolites and re-mapped the fruit metabolites of Wahyuni et al. (2014) on the same version of the linkage map. Data from both sets were log10-transformed before mapping. Potential QTLs for metabolites were identified by interval mapping using the MapQTL 6.0 package (Van Ooijen 2009). Based on permutation tests (1000 iterations) with 10 randomly selected leaf and 10 randomly selected fruit metabolites, common genome-wide LOD thresholds for QTL detection of 3.4 for the leaf metabolites and 3.5 for the fruit metabolites were determined. Metabolite QTLs were considered as co-localized with resistance QTL when there was overlap between the 2-LOD intervals regions of the metabolite and the resistance QTL (Maharijaya et al. 2015).
Correlation analysis of metabolites with thrips resistance parameters
Pearson correlation coefficients of intensity data for all leaf metabolites (log10 transformed data) versus F. occidentalis larval survival. The larval survival data were obtained as described in Maharijaya et al. (2015). In short, per F2 plant five detached leaves were each infested with five L1 larvae; the fraction of survival after 8 days was calculated and transformed as asin(sqrt(x)).
Results
QTL mapping of metabolites in leaf and fruit
The untargeted processing of the LC–MS data of leaves from the F2 population resulted in the detection of 674 centrotypes, hereafter referred to as (putative) metabolites. For 304 leaf metabolites (45% of total), at least one QTL was detected by interval mapping (Supplementary Table 1). For 228 metabolites, a single QTL was found, while for 76 others multiple QTLs were found with a maximum number of 3.
The leaf metabolite QTLs (mQTLs) were spread unevenly over the chromosomes. There were “hotspots” on two linkage groups (P01 and P03), where multiple mQTLs (> 15) co-localized. In contrast, there were also linkage groups with only a few mQTLs (Fig. 1).
Distribution of the number of metabolites that has a significant QTL LOD maximum at each marker, for a the leaf metabolites and b the fruit metabolites (re-mapped data from Wahyuni et al. 2014) in an F2 population of a Capsicum annuum × C. chinense. The 12 chromosomes and six unassigned linkage groups are indicated. The X-axis indicates cumulative genetic positions in cM
Wahyuni et al. (2014) obtained data on metabolites in pepper fruits, in the same F2 population as used in the current study. In order to compare the location of QTLs for fruit metabolites with those of leaf metabolites, we re-mapped the fruit metabolites on the map of Maharijaya et al. (2015), which is an extended version of the map used by Wahyuni et al. (2014). This re-mapping resulted in QTLs for 257 fruit metabolites. The positions of the mQTL hotspots clearly differed between leaves and fruits (Fig. 1) as for fruit mQTLs hotspots were found on Chromosome 9 only.
Metabolites significantly correlating with larval survival of thrips
We used the thrips resistance data of Maharijaya et al. (2015) to calculate the correlations with metabolites. Twenty-three metabolites were significantly correlated (P < 0.01) with larval survival (Table 1), of which 16 were negatively correlated and seven positively. The negatively correlating metabolites were tentatively identified as diterpene glycosides and flavonoids, including a.o. capsianoside IX, luteolin-methyl-O-di-hexose, and Icariside E3, while positively correlating metabolites included a.o. the flavonoid phloretin-C-diglycoside (Table 1).
Co-localization of mQTLs with the QTLs for thrips resistance in pepper
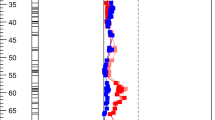
In this F2 population, we previously mapped QTLs for three thrips resistance parameters: survival of the L1 larvae to the L2 stage, to the pre-pupa stage, and leaf damage, all co-localizing on chromosome 6 (linkage group P06) near marker HpmsE078 at position 108 cM Maharijaya et al. (2015). On the same linkage group, we currently detected QTLs for 32 leaf metabolites (Supplementary Table 1). For six of these, the 2-LOD intervals overlapped with those of the resistance QTLs (Fig. 2). Four of these metabolites (LC1980, LC2809, LC5046, and LC5703, see Table 1) were significantly correlated with thrips resistance. Additionally, two metabolites (LC2097 and LC2672) had mQTLs overlapping with the resistance QTL (Fig. 2), but were not significantly correlated with larval survival.
LOD profiles and 1- and 2-LOD QTL intervals for three thrips resistance parameters (black; solid line: damage, dashed: larval survival to L2 stage, dot-dashed: survival to pupa stage; Maharijaya et al. 2015) and of six metabolites whose 2-LOD mQTL interval overlapped with the resistance QTL. Asterisk indicates that the metabolite content also significantly (P < 0.01) correlated with survival to L2 stage. The dotted line at LOD 3.4 indicates the common LOD threshold for the metabolites. These metabolites were tentatively identified: LC1980 as Dihydroxy-megastigmenone hexoside, LC2097 as (+)-lyoniresinol-3-alpha-O-beta-d-glucopyranoside, LC2672 and LC2809 as Icariside E3 or an isomer, LC5046 as a diterpene glycoside, LC5703 as Capsianoside IX
Discussion
QTLs for metabolites in leaves and fruits
We detected 674 semi-polar metabolites in leaves of pepper plants using an untargeted LC–MS-based metabolomics approach. For 45% of them, we could detect mQTLs, which is comparable to other studies [e.g., 43% in pepper fruit (Wahyuni et al. 2014) and 72% in potato tubers (Carreno-Quintero et al. 2012)].
The mQTLs were unevenly spread over the genome, resulting in ‘hotspots’ and ‘coldspots.’ The existence of hot- and coldspots of QTLs for metabolite abundance is a common phenomenon in plants, as shown for example in Arabidopsis thaliana (Keurentjes et al. 2006; Knoch et al. 2017), rice (Matsuda et al. 2012), apple (Khan et al. 2012a), maize (Han et al. 2014), potato (Carreno-Quintero et al. 2012), and also fruits of pepper (Wahyuni et al. 2014). The existence of hotspots is one of the central concepts in genetical genomics, where a single polymorphism leads to widespread changes in the expression of downstream genes, of which the expression therefore maps to the same genomic locus (Breitling et al. 2008). A hotspot for mQTLs suggests the presence of a regulator gene controlling the synthesis or breakdown of a large group of metabolites present at that map position (Saito and Matsuda 2010; Knoch et al. 2017), the clustering of metabolites that are part of the same biosynthetic pathway (Wentzell et al. 2007), a single structural gene encoding a non-specific enzyme that can modify a large series of compounds in a similar manner, e.g., Acyl/Glycosyltransferase (Schwab 2003; Chen et al. 2014), or a single key enzyme in a pathway, e.g., PAL or CHS in case of phenylpropanoids or flavonoids (Khan et al. 2012b). Interestingly, the mQTL hotspot pattern clearly differs between fruits and leaves of pepper with only one major hotspot on chromosome P09 being shared between leaves and fruits. This difference reflects a tissue-specific regulation of the metabolite composition of leaves and fruits of pepper plants. Schauer et al. (2005) also reported a different composition of metabolites in fruit and leaves, in tomato. The mQTL on chromosome P09 that was found in both fruits and leaves affected capsianoside levels in both organs and may encode a regulatory gene which controls the entire capsianoside pathway or a modifying enzyme leading to the production of structurally different capsianosides (Wahyuni et al. 2014).
Metabolite QTLs linked to thrips resistance
In this study, we detected 23 metabolites that were significantly correlated to thrips L1 larval survival (Table 1), which is the life-history parameter that produced the most clear separation among resistant, intermediate, and susceptible accessions (Maharijaya et al. 2012). Although highly significant, the correlations were weak, with R values ranging from of − 0.29 to + 0.25. This may suggest that several metabolites in concert are responsible for the resistance trait observed, rather than a single compound (Van den Oever-Van den Elsen et al. 2016).
Co-localization of QTLs for both thrips resistance and metabolites, as well as the correlation between resistance and specific metabolites may indicate a causal relationship between the two. Of the 23 metabolites that correlated with resistance, 18 could be mapped (Table 1). Four out of the 18 metabolites correlated with thrips resistance co-localized with the major resistance QTL on chromosome 6 (Fig. 2). The abundance of two of these metabolites was negatively correlated with resistance and they were putatively identified as diterpene glycosides (capsianosides), including Capsianoside IX, based on their MSMS data. Interestingly, also the compound that has the strongest negative correlation with thrips development, but could not be mapped, was a diterpene glycoside. So far, there are no other reports that link diterpene glycosides abundance with thrips resistance, though an effect of capsianosides on other insects has been shown previously (Heiling et al. 2010). Further research is needed to confirm the relationship between thrips resistance and diterpene glycoside metabolism. Also, several flavonoids negatively correlate with thrips development (Leiss et al. 2009a). Dihydroxy-megastigmenone hexoside and phloretin-C-diglycoside were positively correlated with resistance, while dihydroxy-megastigmenone hexoside also co-localized with thrips larval development. Both compounds have not been reported in pepper leaves before. Finally, two metabolites (LC2097 and LC2672) co-localized with the resistance QTL while they were not significantly correlated (P > 0.01) to thrips resistance, which may be due to the relatively low percentage of variation explained by these mQTLs, as well as to the fact that one of the metabolites had an additional mQTLs elsewhere in the genome. To confirm causal relationships of metabolites with thrips resistance, it is key to test the candidate compounds in bioassays, and/or to test the effect of candidate genes underlying the mQTLs by silencing or over-expression studies.
In summary, we found only a limited overlap between mQTL hotspots detected in leaves and fruits of pepper. The co-localization of six leaf mQTLs and the previously identified thrips resistance QTL in pepper leaves may indicate a causal relationship. However, the correlation of larval survival with each single metabolite is weak. Further work is still required to confirm the role of correlating and/or co-localizing metabolites, among which diterpene glycosides and flavonoids, in thrips resistance of pepper, or that other yet unknown mechanisms play a larger role. If a causal relationship between (one of) the metabolites and thrips resistance can be shown, the metabolite may be used as a marker for resistance in breeding programs. Also, the compound(s) may be used as leads for identifying the causal gene(s) and for finding resistance in other Capsicum, and perhaps related, species.
References
Abe H, Ohnishi J, Narusaka M, Seo S, Narusaka Y, Tsuda S, Kobayashi M (2008) Function of jasmonate in response and tolerance of Arabidopsis to thrip feeding. Plant Cell Physiol 49(1):68–80
Alonso A, Marsal S, Julià A (2015) Analytical methods in untargeted metabolomics: state of the art in 2015. Front Bioeng Biotechnol 3:23
Blauth SL, Churchill GA, Mutschler MA (1998) Identification of quantitative trait loci associated with acylsugar accumulation using intraspecific populations of the wild tomato, Lycopersicon pennellii. Theor Appl Genet 96(3–4):458–467
Bosland PW, Votava EJ, Votava EM (2012) Peppers: vegetable and spice capsicums, vol 22. CABI, Cambridge
Breitling R, Li Y, Tesson BM, Fu J, Wu C, Wiltshire T, Gerrits A, Bystrykh LV, de Haan G, Su AI, Jansen RC (2008) Genetical genomics: spotlight on QTL hotspots. PLoS Genet 4(10):e1000232
Carreno-Quintero N, Acharjee A, Maliepaard C, Bachem CWB, Mumm R, Bouwmeester H, Visser RGF, Keurentjes JJB (2012) Untargeted metabolic quantitative trait loci analyses reveal a relationship between primary metabolism and potato tuber quality. Plant Physiol 158:1306–1318
ChemnetBase (2013) Directory of natural products. http://dnp.chemnetbase.com. Accessed 2 Feb 2013
Chen W, Gao Y, Xie W, Gong L, Lu K, Wang W, Li Y, Liu X, Zhang H, Dong H, Zhang W, Zhang L, Yu S, Wang G, Lian X, Luo J (2014) Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nature Genet 46:714
De Vos RCH, Moco S, Lommen A, Keurentjes JJB, Bino RJ, Hall RD (2007) Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat Protoc 2(4):778–791
Dunn WB, Erban A, Weber RJ, Creek DJ, Brown M, Breitling R, Hankemeier T, Goodacre R, Neumann S, Kopka J (2013) Mass appeal: metabolite identification in mass spectrometry-focused untargeted metabolomics. Metabolomics 9(1):44–66
FAOSTAT (2016) Food and Agriculture Organization of the United Nations, Statistic Division. http://www.fao.org/faostat/en/. Accessed 26 May 2016
Fery RL, Schalk JM (1991) Resistance in pepper (Capsicum annuum L.) to western flower thrips [Frankliniella occidentalis (Pergande)]. HortScience 26(8):1073–1074
Han Z, Ku L, Zhang Z, Zhang J, Guo S, Liu H, Zhao R, Ren Z, Zhang L, Su H, Dong L, Chen Y (2014) QTLs for seed vigor-related traits identified in maize seeds germinated under artificial aging conditions. PLoS ONE 9(3):e92535
Heiling S, Schuman MC, Schoettner M, Mukerjee P, Berger B, Schneider B, Jassbi AR, Baldwin IT (2010) Jasmonate and ppHsystemin regulate key malonylation steps in the biosynthesis of 17-hydroxygeranyllinalool diterpene glycosides, an abundant and effective direct defense against herbivores in Nicotiana attenuata. Plant Cell 22(1):273
Howard LR, Wildman REC (2007) Antioxidant vitamin and phytochemical content of fresh and processed pepper fruit (Capsicum annuum). In: Wildman REC (ed) Handbook of nutraceutical and functional foods, 2nd edn. CRC Press, Boca Raton, pp 165–192
Keurentjes JJB, Fu J, De Vos CHR, Lommen A, Hall RD, Bino RJ, Van Der Plas LHW, Jansen RC, Vreugdenhil D, Koornneef M (2006) The genetics of plant metabolism. Nat Genet 38(7):842–849
Khan SA, Chibon P-Y, de Vos RCH, Schipper BA, Walraven E, Beekwilder J, van Dijk T, Finkers R, Visser RGF, van de Weg EW, Bovy A, Cestaro A, Velasco R, Jacobsen E, Schouten HJ (2012a) Genetic analysis of metabolites in apple fruits indicates an mQTL hotspot for phenolic compounds on linkage group 16. J Exp Bot 63(8):2895–2908
Khan SA, Schaart JG, Beekwilder J, Allan AC, Tikunov YM, Jacobsen E, Schouten HJ (2012b) The mQTL hotspot on linkage group 16 for phenolic compounds in apple fruits is probably the result of a leucoanthocyanidin reductase gene at that locus. BMC Res Notes 5(1):618
Kiers E, De Kogel W, Balkema-Boomstra A, Mollema C (2000) Flower visitation and oviposition behavior of Frankliniella occidentalis (Tysan., Thripidae) on cucumber plants. J Appl Entomol 124(1):27–32
Knoch D, Riewe D, Meyer RC, Boudichevskaia A, Schmidt R, Altmann T (2017) Genetic dissection of metabolite variation in Arabidopsis seeds: evidence for mQTL hotspots and a master regulatory locus of seed metabolism. J Exp Bot 68(7):1655–1667
Leiss KA, Choi YH, Abdel-Farid IB, Verpoorte R, Klinkhamer PGL (2009a) NMR metabolomics of thrips (Frankliniella occidentalis) resistance in senecio hybrids. J Chem Ecol 35(2):219–229
Leiss KA, Maltese F, Choi YH, Verpoorte R, Klinkhamer PGL (2009b) Identification of chlorogenic acid as a resistance factor for thrips in chrysanthemum. Plant Physiol 150(3):1567–1575
Lommen A (2009) MetAlign: interface-driven, versatile metabolomics tool for hyphenated full-scan mass spectrometry data preprocessing. Anal Chem 81(8):3079–3086
Maharijaya A, Vosman B, Steenhuis-Broers G, Harpenas A, Purwito A, Visser RGF, Voorrips RE (2011) Screening of pepper accessions for resistance against two thrips species (Frankliniella occidentalis and Thrips parvispinus). Euphytica 177(3):401–410
Maharijaya A, Vosman B, Verstappen F, Steenhuis-Broers G, Mumm R, Purwito A, Visser RG, Voorrips RE (2012) Resistance factors in pepper inhibit larval development of thrips (Frankliniella occidentalis). Entomol Exp Appl 145(1):62–71
Maharijaya A, Vosman B, Steenhuis-Broers G, Pelgrom K, Purwito A, Visser RF, Voorrips R (2015) QTL mapping of thrips resistance in pepper. Theor Appl Genet 128(10):1945–1956
Marin A, Ferreres F, Tomas-Barberan FA, Gil MI (2004) Characterization and quantitation of antioxidant constituents of sweet pepper (Capsicum annuum L.). J Agric Food Chem 52(12):3861–3869
Maris PC, Joosten NN, Goldbach RW, Peters D (2003) Restricted spread of Tomato spotted wilt virus in thrips-resistant pepper. Phytopathol 93:1223–1227
Matsuda F, Okazaki Y, Oikawa A, Kusano M, Nakabayashi R, Kikuchi J, Yonemaru J-I, Ebana K, Yano M, Saito K (2012) Dissection of genotype–phenotype associations in rice grains using metabolome quantitative trait loci analysis. Plant J 70(4):624–636
Miller DS (2001) Review of oleoresin capsicum (pepper) sprays for self-defense against captive wildlife. Zoo Biol 20(5):389–398
Nakamura Y, Asahi H, Altaf-Ul-Amin M, Kurokawa K, Kanaya S (2008) KNApSAcK: A comprehensive species-metabolite relationship database. Accessed 2 Feb 2013
Saito K, Matsuda F (2010) Metabolomics for functional genomics, systems biology, and biotechnology. Annu Rev Plant Biol 61(1):463–489
Schauer N, Zamir D, Fernie AR (2005) Metabolic profiling of leaves and fruit of wild species tomato: a survey of the Solanum lycopersicum complex. J Exp Bot 56(410):297–307
Schwab W (2003) Metabolome diversity: too few genes, too many metabolites? Phytochem 62(6):837–849
Smith CM (2005) Plant resistance to arthropods: molecular and conventional approaches. Springer Science & Business Media, Dordrecht, pp 65–99
Stummel RJ, Bosland PW (2007) Ornamental pepper. In: Anderson NO (ed) Flower breeding and genetics. Springer, Netherlands, pp 561–599
Tikunov Y, Lommen A, de Vos CHR, Verhoeven HA, Bino RJ, Hall RD, Bovy AG (2005) A novel approach for nontargeted data analysis for metabolomics. Large-scale profiling of tomato fruit volatiles. Plant Physiol 139(3):1125–1137
Tikunov YM, Laptenok S, Hall RD, Bovy A, Vos RCH (2012) MSClust: a tool for unsupervised mass spectra extraction of chromatography-mass spectrometry ion-wise aligned data. Metabolomics 8(4):714–718
Topuz A, Ozdemir F (2007) Assessment of carotenoids, capsaicinoids and ascorbic acid composition of some selected pepper cultivars (Capsicum annuum L.) grown in Turkey. J Food Compos Anal 20(7):596–602
Trichilo PJ, Leigh TF (1988) Influence of resource quality on the reproductive fitness of flower thrips (Thysanoptera: Thripidae). Ann Entomol Soc Am 81(1):64–70
Ullman DE, Meideros R, Campbell LR, Whitfield AE, Sherwood JL, German TL (2002) Thrips as vectors of tospoviruses. In: Kader JC, Delseny M (eds) Advances in botanical research, vol 36. Academic Press, Boston, pp 113–140
Van Ooijen JW (2009) MapQTL6, software for the mapping of quantitative trait loci in experimental population of diploid species. Kyazma BV, Wageningen
Van den Oever-van den Elsen F, Lucatti AF, van Heusden S, Broekgaarden C, Mumm R, Dicke M, Vosman B (2016) Quantitative resistance against Bemisia tabaci in Solanum pennellii: genetics and metabolomics. J Integr Plant Biol 58 (4):397–412
Vinaixa M, Schymanski EL, Neumann S, Navarro M, Salek RM, Yanes O (2016) Mass spectral databases for LC/MS- and GC/MS-based metabolomics: state of the field and future prospects. Trends Anal Chem 78:23–35
Vosman B, Van’t Westende W, Henken B, Van Eekelen H, De Vos R, Voorrips RE (2018) Broad spectrum insect resistance and metabolites in close relatives of the cultivated tomato. Euphytica 214:46
Wahyuni Y, Ballester AR, Sudarmonowati E, Bino RJ, Bovy AG (2011) Metabolite biodiversity in pepper (Capsicum) fruits of thirty-two diverse accessions: variation in health-related compounds and implications for breeding. Phytochemistry 72(11–12):1358–1370
Wahyuni Y, Ballester AR, Sudarmonowati E, Bino RJ, Bovy AG (2013a) Secondary metabolites of Capsicum species and their importance in the human diet. J Nat Prod 76(4):783–793
Wahyuni Y, Ballester AR, Tikunov Y, De Vos CHR, Pelgrom K, Maharijaya A, Sudarmonowati E, Bino RJ, Bovy AG (2013b) Metabolomics and molecular marker analysis to explore pepper (Capsicum sp.) biodiversity. Metabolomics 9(1):130–144
Wahyuni Y, Stahl-Hermes V, Ballester AR, de Vos RCH, Voorrips RE, Maharijaya A, Molthoff J, Zamora MV, Sudarmonowati E, Arisi ACM, Bino RJ, Bovy AG (2014) Genetic mapping of semi-polar metabolites in pepper fruits (Capsicum sp.): towards unravelling the molecular regulation of flavonoid quantitative trait loci. Mol Breed 33(3):503–518
Wentzell AM, Rowe HC, Hansen BG, Ticconi C, Halkier BA, Kliebenstein DJ (2007) Linking metabolic QTLs with network and cis-eQTLs controlling biosynthetic pathways. PLoS Genet 3(9):e162
Wink M (1988) Plant breeding: importance of plant secondary metabolites for protection against pathogens and herbivores. Theor Appl Genet 75(2):225–233
Acknowledgements
The research was financially supported by the Royal Netherlands Academy of Arts and Sciences in the framework of the Scientific Programme Indonesia—the Netherlands (SPIN), as well as by the Netherlands Metabolomics Centre (RCHdV) and the Centre for BioSystems Genomics (BV, REV, RCHdV) which are both part of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Rupesh Kariyat.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Maharijaya, A., Vosman, B., Pelgrom, K. et al. Genetic variation in phytochemicals in leaves of pepper (Capsicum) in relation to thrips resistance. Arthropod-Plant Interactions 13, 1–9 (2019). https://doi.org/10.1007/s11829-018-9628-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11829-018-9628-7