Abstract
A species’ genotype can have extended consequences for the structure of the surrounding community, but few studies have investigated the extended consequences of genetic variation in animals. Accordingly, I examined the importance of genetically based variation among five populations of the ant-tended aphid Aphis asclepiadis for its interactions with both ants and predators. In a common environment, aphid source population accounted for 23 and 17% of the variation in the occurrence of ants and predators, respectively. Ant exclusion increased predator abundance, accounting for 25% of variation, but there was no detectable influence of ants on aphid abundance. There was an indication that aphid source populations varied in honeydew quality, but this was uncorrelated with rates of ant attendance. This study provides the first evidence for genetic variation in aphids for attractiveness to ants, and underscores the important link between intra-specific genetic variation in aphids and the processes governing arthropod community structure.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A growing number of studies have documented how the structure of ecological communities can be influenced by intra-specific variation in the component species (Agrawal 2003; Johnson and Stinchcombe 2007; Whitham et al. 2006). Most of this work has studied plants, where genotype identity can explain much of the variation in arthropod species composition (Crutsinger et al. 2009; Johnson and Agrawal 2005; Wimp and Whitham 2001) and can mediate the interactions among resident arthropod species (Gassmann and Hare 2005; Johnson 2008; Mooney and Agrawal 2008). Because such extended phenotypes appear to be common, there is an emerging view that intra-specific variation can have important consequences for fundamental aspects of community structure.
Only a handful of studies have investigated the consequences of intra-specific genetic variation in animals for the structure of the communities in which those animals reside. In fish, divergent phenotypes of sticklebacks (Harmon et al. 2009) and alewife (Palkovacs and Post 2008; Post and Palkovacs 2009) can influence prey communities and ecosystem function. For insects, several studies have documented genetic variation in aphid resistance to parasitoids or pathogens in a laboratory setting (Ferrari and Godfray 2006; Ferrari et al. 2001), and one study has tested for genetic variation in enemy recruitment in the field; Hazell and Fellowes (2009) showed that genotypes of pea aphids differentially attracted a predator (larval hoverflies) and a parasitoid. In addition, Tetard-Jones et al. (2007) showed that aphid genotypes differed in their effects on host plant growth. Thus, while evidence for plant genetic effects on community structure is substantial, far less is known about the extent to which parallel dynamics may operate in other trophic interactions.
Genetic variation in the effects of plants or animals on community structure can occur at varying scales. Variation within a population should contribute to variation in community structure over small spatial scales, with temporal changes in community structure being driven by population changes in the genotype frequency across generations. In contrast, partitioning of genetic variation among populations should increase the spatial scale of turnover in community structure, with changes in community structure over time being driven by population differentiation and meta-population dynamics. Consequently, the scale of genetic variation has important implications for both the spatial pattern of variation in community structure, and the interplay between population and meta-population dynamics and community structure. Past studies on both plants and animals have taken various approaches with regards to the scale of genetic variation investigated, ranging from those that compare genotypes (Crutsinger et al. 2006; Ferrari and Godfray 2006; Ferrari et al. 2001; Hazell and Fellowes 2009; Johnson and Agrawal 2005; Mooney and Agrawal 2008; Tetard-Jones et al. 2007) to those that compare distinct populations (Harmon et al. 2009; Madritch et al. 2006; Palkovacs and Post 2008; Post and Palkovacs 2009; Wimp and Whitham 2001; Wooley et al. 2007).
Mutualisms between ants and aphids are a common component of most terrestrial communities and have served as a model for the mutualisms generally (Stadler and Dixon 2008). In these interactions, aphids feeding upon nutrient-poor phloem sap dispose of large quantities of sugar-rich fluid referred to as honeydew. In exchange for honeydew, ants provide a variety of services including protection from predators. Although all aphids produce honeydew, only 40% of aphid species are tended by ants (Stadler 1997). Because many aphid genera include both tended and untended species (Mooney 2006; Mooney et al. 2008; Shingleton and Stern 2003), the aphid traits related to mutualism with ants may be evolutionarily labile.
I report here on a field experiment testing for genetic variation among five populations of the ant-tended aphid Aphis asclepiadis Fitch for its interactions with both ants and predators. I also investigated whether variation among aphid populations for interactions with ants might be due to the attractiveness of honeydew to ants. By investigating such variation among populations, this study explicitly tests for landscape scale variation in aphid genotypes of significance to the associated arthropod community.
Methods
Natural history
The common milkweed (Asclepias syriaca L.) is a widespread, native perennial that occurs throughout eastern North America where it is fed upon by the aphid A. asclepiadis. Aphis asclepiadis, like many aphid species, is cyclically parthenogenetic; it has one sexual generation each autumn and overwinters as an egg, after which the hatching female (fundatrix) and subsequent summer generations give birth to genetically identical live young. In environmental growth chambers set to 25°C, 14 h days and 8 h nights A. asclepiadis individuals reach reproductive maturity at 6 days and have a reproductive lifespan of 15 days over which time they produce nearly 40 nymphs (Mooney et al. 2008).
Aphids were collected from five abandoned agriculture fields near Ithaca, NY USA, with one of these fields being the site for a common garden experiments. In each field, milkweed is fed upon by A. asclepiadis and tended by the ant Formica podzolica Francour. Past work has demonstrated that F. podzolica can have positive effects on A. asclepiadis abundance, but that such effects can depend on several ecological factors including milkweed genotype (Mooney and Agrawal 2008) and whether or not A. asclepiadis is in competition with other aphid species (Smith et al. 2008).
Experimental protocols
Aphids were collected from five populations in abandoned agricultural fields arranged in a transect with Neimi as the northernmost site, and progressing southward to Whipple Farm, Monkey Run, Ellis Hollow, and ending with Dunlop 13 km south of Neimi. Four aphid genotypes (the collection of aphids found on one milkweed stem) separated by a minimum of 25 m were collected from each site in early June 2007, shortly after egg hatch. These aphid colonies were likely to be genetically distinct because they each consisted of a fundatrix and a small number of nymphs. But because genetic variation was not measured at the molecular level, the failure to detect ecological variation among aphid populations might be due to a lack of genetic differentiation. Nevertheless, the fact that differences were seen in a common setting (see below) demonstrates that ecologically relevant genetic variance was present among these sites.
Several steps were taken to control for aphid maternal effects and for host plant effects on aphid traits. After collection, the aphids of each genotype were split and placed on three potted and individually caged A. syriaca seedlings in an environmental growth chamber maintained at a constant 25°C with 14-h days and 10-h nights. Plants were grown from genetically variable seed collected in the Ithaca area, had between 4 and 8 true leaves, were watered as needed and fertilized weekly. On 25–26 June, after approximately two aphid generations, two adult aphids from each genotype were in turn transferred to each of seven new seedlings for 48 h to produce two nymphs, after which the adults were removed.
Honeydew assay
The original three seedlings hosting each aphid genotype in the environmental growth chambers were used to collect honeydew samples. From July 7–8, two pre-weighed 1.0 cm2 aluminum foil disks were placed beneath aphids on each plant for two days to collect honeydew. The two foil disks on each plant were placed on opposing sides of the plant. Theses collections yielded six honeydew-coated foil disks for each aphid genotype. After weighing each disk again to determine the initial honeydew mass, six honeydew arrays were constructed by affixing one foil disk from each aphid genotype to a cardboard sheet with a pin, randomizing the location of the 20 foil disks on each array.
On July 9, six F. podzolica mounds were selected at the Neimi site. Each honeydew array was placed 0.5 m away from a mound and left for 15 min, after which time the number of ants present on each foil disk was counted. The six arrays were then rotated to new ant mounds such that at the end of six trials each array had been placed adjacent to each ant mound. At the conclusion of the trials, each foil disk was again weighed to determine the amount of honeydew removed by ants. These measurements thus produced values for initial honeydew mass and honeydew mass removed by ants for each foil disk. Because the honeydew offered to ants was collected the day prior to this bioassay, its water content and other aspects of its composition almost certainly was altered as compared to what the aphids would present directly to ants. Consequently, this bioassay assesses only the relative attractiveness of aphid honeydew to ants. At the same time, ants also feed from honeydew accumulations on leaf surfaces (Mooney and Agrawal 2008; Morales 2000), a situation analogous to that used in this bioassay.
Aphid assay
On July 7, the seven potted seedlings from each of the 20 aphid genotypes were placed around four F. podzolica mounds at the Neimi site. At that time, the 140 plants averaged 8 ± 0.8 (mean ± SE) aphids each. Half of the seedlings were placed in an ant exclusion treatment in which the pot exterior was coated with Insect-a-Slip Insect Barrier (Bioquip, Rancho Dominguez, CA USA). Replicates were randomized with respect to ant mound and ant exclusion such that all genotypes and ant mounds had plants apportioned approximately equally between ant exclusion and control treatments. Aphid, ant and predator abundances were counted on each plant on 7 July, 9 July, 12 July and 14 July. On each date, aphid abundance was taken once while three consecutive counts were made over 90 min for relatively mobile ants and predators and then averaged. While the ant exclusion treatment might also prevent access from unwinged aphid predators, the two types of predators observed—adult Coccinellidae (Coleoptera) and Miridae (Hemiptera)—are both winged and thus not directly influenced by this manipulation.
Statistical analyses
This experiment was originally designed to test for genetic effects of aphids on ants and predators in terms of both population- and genotype-level variation. In preliminary analyses, response variables were modeled as a function of aphid genotype nested within source population. Unfortunately, the distributions of these data were non-normal, no adequate transformation to normalize these data was found, and the outcomes of analyses based upon generalized linear models that can accommodate non-normal data were highly contingent upon model assumptions. It was thus impossible to reach any reliable conclusions about variation among individual aphid genotypes. In contrast, dependent variables based upon genotype means (pooling the data from replicates of a genotype) were all normally distributed and could thus provide reliable tests for genetically based variation among aphid populations. Consequently, all analyses test for effects of aphid source population, with replication coming from four genotype means per population.
For the honeydew assay, aphid genotype means were calculated for the number of ants observed across all honeydew arrays at all ant mounds, as well as for the original mass of honeydew present on the six foil disks. Ant abundance was then modeled as a function of honeydew mass and aphid source population using analysis of covariance (ANCOVA).
For the aphid assay, aphid abundance increased over time, counts were highly correlated among repeated measures, and ant abundance was highly correlated with aphid abundance at each time point; consequently, only the 14 July counts of aphids and ants were used in analyses. In contrast, counts of predators were extremely low at each time point and analyses were based upon total predator abundance observed across all sampling dates.
Least square means for aphid and ant abundance were calculated for each aphid genotype in ant exclusion and control treatments, thus removing variation due to ant mound. Analyses were then performed with these aphid genotype means as the dependent variables. Aphid population growth, defined here as aphids*initial aphid−1*day−1, was modeled as a function of ant exclusion, aphid source population and their interaction using analysis of variance (ANOVA). Ant abundance (on seedlings with ant access) was modeled as a function of aphid source population, aphid abundance and their interaction using ANCOVA. In both analyses, non-significant (P > 0.15) interaction terms were removed.
Predator data were analyzed in two complementary ways: First, each aphid genotype was scored for the proportion of replicate plants on which one or more predators were observed, and this was done separately for treatments with and without ants. These arcsine-square root transformed proportions were then modeled as a function of ant exclusion, aphid source population and their interaction using ANOVA. This analysis revealed that the effects of aphid source population on predators did not depend on the presence of ants (see below). Consequently, a second analytical approach was taken in which aphid source populations were compared across both levels of the ant exclusion treatment. Here, each genotype was scored for whether or not predators were observed on any of the seven replicate host plants. Aphid source populations were then compared with respect to the number of aphid genotypes scored as having predators, or not, using a Fisher’s exact test.
Fitness costs for ant recruitment, predator resistance and honeydew attractiveness were tested for by correlating source population means for each variable with the mean population growth rate for that source population. Each correlation was based upon aphid source population means and thus had a relatively small sample size (N = 5).
Results
Honeydew assay
The original mass of honeydew had a positive effect on the number of ants attending each foil disk (F 1,14 = 9.86, P = 0.0072). Statistically adjusting for original honeydew mass, there was a trend towards aphid source populations differing in the attractiveness of their honeydew (F 4,19 = 2.68, P = 0.08, R 2 = 0.31; Fig. 1a).
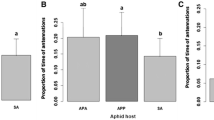
Variation among aphid source populations for aphid honeydew attractiveness to ants (a), aphid population growth rate (b), ant recruitment to aphids (c) and predator recruitment to aphids (d). Aphid source populations are coded with letters (n Neimi, w Whipple Farm, m Monkey Run, e Ellis Hollow, d Dunlop). Values in a–c are based upon source population means (±1 SE) of four aphid genotypes each, and P values for source population effects come from ANOVA. Values in d are the proportions of aphid genotypes (N = 4) from which one or more predators were observed on any of seven replicate host plants, and P values for source population effects come from a Fisher’s exact test
Aphid assay
Aphid population growth rate was not significantly influenced by aphid source population (F 4,15 = 1.80, P = 0.18; Fig. 1b) or the presence of ants (F 1,19 = 0.69, P = 0.42; Fig. 2) in a reduced model that excluded the non-significant interaction term (F 4,15 = 0.87, P = 0.51).
Ant abundance was influenced by both aphid abundance (F 4,19 = 26.89, P < 0.0001) and aphid source population (F 4,19 = 3.32, P = 0.0415, R 2 = 0.23) in a reduced statistical model that excluded the non-significant interaction term (F 4,19 = 0.41, P = 0.80). Ant abundance varied more than two-fold among aphid source populations after controlling for aphid abundance (Fig. 1c).
The predators observed in association with aphids consisted of adult Coccinellidae (Coleoptera) and predatory Miridae (Hemiptera). Because of lower predator abundance, these taxonomically distinct groups were pooled and only total predator abundance was analyzed. There was a trend towards a significant effect of aphid source population on predator abundance, quantified as the proportion of plants with one or more predators (F 4,15 = 2.55, P = 0.08, R 2=0.17; Figure S1) and ants had a strong negative effect (F 1,19 = 14.58, P = 0.0012, R 2 = 0.25; Fig. 2) in a reduced statistical model that excluded the non-significant interaction term (F 4,15 = 1.64, P = 0.22). At the same time, a Fisher’s exact test showed an effect of source population on predators when predator abundance was quantified as the number of aphid genotypes for which predators were or were not observed on any of the seven host plants (P = 0.02; Fig. 1d). Predator abundance varied dramatically among aphid source populations, ranging from Ellis Hollow aphids, which attracted no predators, to those from Dunlop, which attracted predators to all aphid clones (Fig. 1d) and to nearly one-third of replicate host plants (Figure S1).
Trade-offs
There was no detectable relationship among the attractiveness of an aphid source population’s honeydew, the rate at which those aphids were tended by ants, predator recruitment and the growth rate of aphid source populations (P ≥ 0.25 for all pairwise tests).
Discussion
Aphid source population explained nearly 23 and 17% of the variation in ant and predator abundance, respectively, effects that were comparable to the 25% of variation in predator abundance attributable to the effects of ants. Ant-aphid mutualisms are keystone interactions because aphid-recruited ants can have far-reaching effects on both arthropod communities and plants (Styrsky and Eubanks 2007). Along these lines, past studies have shown how ant-aphid interactions can vary strongly among milkweed genotypes, and that milkweed genetic effects on ant-aphid interactions can in turn influence the survival of other milkweed herbivores (Mooney and Agrawal 2008). As a consequence, genetic variation among aphids might be expected to influence other arthropods not only directly (Hazell and Fellowes 2009), but also indirectly via direct effects on ant abundance.
Molecular markers were not used in this study, and there is no direct evidence for the level of genetic differentiation among the genotypes and populations studied here. Because the experimental design established a common environment and controlled for maternal effects, the observed variation among populations provides strong indirect evidence for genetic differentiation. Less certain is whether collections of multiple aphid clones within populations constitute separate genotypes. While there is no direct evidence on this mater, each genotype is likely unique because aphid clones were collected shortly after emergence from the over-wintering, sexually produced egg stage. Under the unlikely circumstances that replicate aphid clones from a site were of a single genotype, these results would constitute evidence for genetic variation at the genotypic instead of population level. Consequently, the results presented here provide strong evidence for an aphid genetic influence on ants and predators, with the level of genetic variation (genotypic or population) being somewhat less certain.
The aphid traits responsible for influencing ant recruitment are unclear. There was evidence that aphid source populations varied in honeydew attractiveness (Fig. 1a). Yet this variation was unrelated to ant attendance of aphids (Fig. 1c), perhaps because the bioassay used honeydew that was 24–48 h old. At the same time, this bioassay was ecologically realistic in that ants often collect accumulated honeydew directly from leaves (Mooney and Agrawal 2008; Morales 2000). Ant attendance is also expected to vary in accordance with aphid abundance, but aphid source populations did not vary significantly in population size (Fig. 1b) and there was no detectable genetic correlation between aphid source population means for aphid and ant abundance (Fig. 1b, c). Past work comparing tended and untended aphid species demonstrates a number of features associated with ant mutualism, including longer mouthparts (Oliver et al. 2008; Shingleton and Stern 2003; Shingleton et al. 2005), shorter cornicles (Mondor et al. 2002) and winglessness (Stadler et al. 2003). Yet each of these adaptations is more likely associated with a loss of predator avoidance traits than with an increase in attractiveness to ants. It may be that the genetic variation in ant recruitment found here is due to variation in the quantity of honeydew production and perhaps to variation in behavior or morphology associated with how honeydew is presented to ants.
Genetic variation among aphid populations for predator abundance was apparently a direct effect of aphids on predators (Hazell and Fellowes 2009), and was not mediated by differential ant recruitment. While ants negatively influenced predators (Fig. 2), there was no genetic correlation between ant and predator abundance among the aphid source populations (Fig. 1c, d). Consequently, aphid genetic variation for predator abundance was likely due to some form of direct influence as has previously been shown by Hazell and Fellowes (2009). Plausible explanations include variation among aphid source populations in predator-attracting volatile emissions, or perhaps variation in aphid sequestration of plant secondary metabolites (Mooney et al. 2008).
Even though ants strongly reduced predator abundance, they did not provide a beneficial effect to aphids in this study. Two past studies at this same field site have found beneficial effects of F. podzolica for A. asclepiadis, but only under certain circumstances. Smith et al. (2008) suggested that the principal benefit of ants may not be protection from predators, but rather ants may reduce competition for tended aphids by reducing the abundance of untended aphid species. In the present experiment, herbivores besides A. asclepiadis were absent, perhaps making it unlikely that ants would provide a detectable benefit to this aphid via competitive release. In addition, Mooney and Agrawal (2008) showed ants increase aphid abundance on some milkweed genotypes but have negative effects on others. While the seedlings used in this experiment were genetically variable, it is nonetheless possible that a benefit of ants would have been detected if we had used a different seed source for plants.
Studies investigating the consequences for genetic variation have either compared genotypes co-occurring in a single population (e.g. Crutsinger et al. 2006; Ferrari and Godfray 2006; Ferrari et al. 2001; Hazell and Fellowes 2009; Johnson and Agrawal 2005; Mooney and Agrawal 2008) or individual genotypes from distinct populations but without population replication (e.g. Madritch et al. 2006; Wimp and Whitham 2001; Wooley et al. 2007). Noticeably lacking are studies that explicitly test for intra-specific genetic variation at the population level. While there are examples of local adaptation leading to population differences (Harmon et al. 2009; Palkovacs and Post 2008; Post and Palkovacs 2009), such cases stem from evolutionary divergence among populations isolated for thousands of years. It is thus notable that the present study documents differences among closely situated populations, despite the likelihood of aphid movement and gene flow.
This study contributes to a growing body of empirical work that demonstrates the community-wide consequences of intra-specific genetic variation. A number of past studies have documented the importance of plant genotype identity for arthropod community structure (Crutsinger et al. 2009; Gassmann and Hare 2005; Johnson 2008; Johnson and Agrawal 2005; Mooney and Agrawal 2008; Wimp and Whitham 2001). This study provides important additional evidence that such effects can similarly be attributed to animals generally (Ferrari and Godfray 2006; Ferrari et al. 2001; Harmon et al. 2009; Hazell and Fellowes 2009; Palkovacs and Post 2008; Post and Palkovacs 2009), and provides the first documentation of such effects on ant-aphid interactions. Furthermore, the proportion of variation in arthropod community structure associated with aphid genetic variation was comparable to that of ant exclusion. Consequently, these results place the influence of aphid genotype on a par with other factors generally considered to be of high ecological relevance.
References
Agrawal AA (2003) Community genetics: new insights into community ecology by integrating population genetics. Ecology 84:543–544
Crutsinger GM, Collins MD, Fordyce JA, Gompert Z, Nice CC, Sanders NJ (2006) Plant genotypic diversity predicts community structure and governs an ecosystem process. Science 313:966–968
Crutsinger GM, Cadotte MW, Sanders NJ (2009) Plant genetics shapes inquiline community structure across spatial scales. Ecol Lett 12:285–292
Ferrari J, Godfray HCJ (2006) The maintenance of intraspecific biodiversity: the interplay of selection on resource use and on natural enemy resistance in the pea aphid. Ecol Res 21:9–16
Ferrari J, Muller CB, Kraaijeveld AR, Godfray HCJ (2001) Clonal variation and covariation in aphid resistance to parasitoids and a pathogen. Evolution 55:1805–1814
Gassmann AJ, Hare JD (2005) Indirect cost of a defensive trait: variation in trichome type affects the natural enemies of herbivorous insects on Datura wrightii. Oecologia 144:62–71
Harmon LJ, Matthews B, Des Roches S, Chase JM, Shurin JB, Schluter D (2009) Evolutionary diversification in stickleback affects ecosystem functioning. Nature 458:1167–1170
Hazell SP, Fellowes MDE (2009) Intra-specific variation affects the structure of the natural enemy assemblage attacking pea aphid colonies. Ecol Entomol 34:34–42
Johnson MTJ (2008) Bottom-up effects of plant genotype on aphids, ants, and predators. Ecology 89:145–154
Johnson MTJ, Agrawal AA (2005) Plant genotype and environment interact to shape a diverse arthropod community on evening primrose (Oenothera biennis). Ecology 86:874–885
Johnson MTJ, Stinchcombe JR (2007) An emerging synthesis between community ecology and evolutionary biology. Trends Ecol Evol 22:250–257
Madritch M, Donaldson JR, Lindroth RL (2006) Genetic identity of Populus tremuloides litter influences decomposition and nutrient release in a mixed forest stand. Ecosystems 9:528–537
Mondor EB, Roitberg BD, Stadler B (2002) Cornicle length in Macrosiphini aphids: a comparison of ecological traits. Ecol Entomol 27:758–762
Mooney KA (2006) The disruption of an ant–aphid mutualism increases the effects of birds on pine herbivores. Ecology 87:1805–1815
Mooney KA, Agrawal AA (2008) Plant genotype shapes ant–aphid interactions: implications for community structure and indirect plant defense. Am Nat 168:E195–E205
Mooney KA, Jones P, Agrawal AA (2008) Coexisting congeners: demography, competition, and interactions with cardenolides for two milkweed-feeding aphids. Oikos 117:450–458
Morales MA (2000) Mechanisms and density dependence of benefit in an ant-membracid mutualism. Ecology 81:482–489
Oliver TH, Leather SR, Cook JM (2008) Macroevolutionary patterns in the origin of mutualisms involving ants. J Evol Biol 21:1597–1608
Palkovacs EP, Post DM (2008) Eco-evolutionary interactions between predators and prey: can predator-induced changes to prey communities feed back to shape predator foraging traits? Evol Ecol Res 10:699–720
Post DM, Palkovacs EP (2009) Eco-evolutionary feedbacks in community and ecosystem ecology: interactions between the ecological theatre and the evolutionary play. Phil Trans Royal Soc B-Biol Sci 364:1629–1640
Shingleton AW, Stern DL (2003) Molecular phylogenetic evidence for multiple gains or losses of ant mutualism within the aphid genus Chaitophorus. Mol Phylogenet Evol 26:26–35
Shingleton AW, Stern DL, Foster WA (2005) The origin of a mutualism: a morphological trait promoting the evolution of ant-aphid mutualisms. Evolution 59:921–926
Smith RA, Mooney KA, Agrawal AA (2008) Coexistence of three specialist aphids on common milkweed, Asclepias syriaca. Ecology 89:2187–2196
Stadler B (1997) The relative importance of host plants, natural enemies and ants in the evolution of life-history characters in aphids. In: Dettner K, Bauer G, Vèolkl W (eds) Vertical food web interactions: evolutionary patterns and driving forces. Springer, Berlin; New York, pp 241–256
Stadler B, Dixon AFG (1998) Costs of ant attendance for aphids. J Animal Ecol 67:454–459
Stadler B, Dixon AFG (2008) Mutualism: ants and their insect partners. Cambridge University Press, Cambridge; New York
Stadler B, Dixon AFG, Kindlmann P (2002) Relative fitness of aphids: effects of plant quality and ants. Ecol Lett 5:216–222
Stadler B, Kindlmann P, Smilauer P, Fiedler K (2003) A comparative analysis of morphological and ecological characters of European aphids and lycaenids in relation to ant attendance. Oecologia 135:422–430
Styrsky JD, Eubanks MD (2007) Ecological consequences of interactions between ants and honeydew-producing insects. Proc Royal Soc B-Biol Sci 274:151–164
Tetard-Jones C, Kertesz MA, Gallois P, Preziosi RF (2007) Genotype-by-genotype interactions modified by a third species in a plant-insect system. Am Nat 170:492–499
Whitham TG et al (2006) A framework for community and ecosystem genetics: from genes to ecosystems. Nat Rev Genet 7:510–523
Wimp GM, Whitham TG (2001) Biodiversity consequences of predation and host plant hybridization on an aphid–ant mutualism. Ecology 82:440–452
Wooley SC, Donaldson JR, Gusse AC, Lindroth RL, Stevens MT (2007) Extrafloral nectaries in aspen (Populus tremuloides): heritable genetic variation and herbivore-induced expression. Ann Bot 100:1337–1346
Acknowledgments
This work was supported by the NSF-DEB 0919178 and the School of Biological Sciences at the University of California at Irvine. R. Alex Smith provided assistance in the field and Anurag Agrawal, Marc Johnson and two anonymous reviewers provided useful comments on this manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Jan Pettersson.
Electronic supplementary material
Below is the link to the electronic supplementary material.

11829_2010_9110_MOESM1_ESM.jpg
Figure S1. Aphid source population means (±1 SE) for predator recruitment to aphids. Aphid source populations are coded with letters (n=Neimi, w=Whipple Farm, m=Monkey Run, e=Ellis Hollow, d=Dunlop). Predator recruitment for each aphid clone is scored as the proportion of plants on which one or more predators were observed, and source population means are then based upon the mean of four aphid genotypes. P values for aphid source population effects from ANOVA are provided at the top of each panel. Supplementary material 1 (jpg 20 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Mooney, K.A. Genetically based population variation in aphid association with ants and predators. Arthropod-Plant Interactions 5, 1–7 (2011). https://doi.org/10.1007/s11829-010-9110-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11829-010-9110-7