Abstract
Sanger sequencing revolutionized molecular genetics 40 years ago. However, next-generation sequencing technologies became further game changers and shaped our current view on genome structure and function in health and disease. Although still at the very beginning, third-generation sequencing methods, also referred to as long-read sequencing technologies, provide exciting possibilities for studying structural variations, epigenetic modifications, or repetitive elements and complex regions of the genome. We discuss the advantages and pitfalls of current long-read sequencing methods with a focus on nanopore sequencing, summarize respective applications and provide an outlook on the potential of these novel methods.
Zusammenfassung
Nachdem die Sanger-Sequenzierung vor vierzig Jahren die Lebenswissenschaften revolutioniert hat, prägen die Next-Generation-Sequencing-Technologien unsere derzeitige Sicht auf die Genomik. Dies gilt sowohl für das Verständnis von Genomaufbau und -funktion als auch für die Erforschung und Diagnostik von Erkrankungen. Durch die jüngsten Verfahren des „third-generation sequencing“, auch als „long-read sequencing“ bezeichnet, ergeben sich weitreichende Möglichkeiten, strukturelle Varianten, epigenetische Modifikationen oder repetitive Elemente und komplexe Regionen des Genoms im Detail zu untersuchen. Der Artikel gibt eine Übersicht über Vor- und Nachteile aktueller Long-Read-Sequencing-Verfahren mit einem Schwerpunkt im Bereich der Nanoporensequenzierung und fasst deren Potenzial und Anwendungsmöglichkeiten zusammen.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Initial sequencing technologies and short-read next-generation sequencing
Determining the nucleic acid sequence has shaped our view of genome structure and function. Back in 1968, Wu and Kaiser used primer extension methods to identify a short sequence of the bacteriophage lambda [62], whereas 5 years later, Maxam and Gilbert determined the sequence of the lactose-repressor binding site by chemical cleavage [21]. Subsequently, the widespread method using chain-terminating dideoxynucleotides by Frederick Sanger and colleagues has fostered sequencing since the mid-1970s [42, 51]. Sanger sequencing culminated in the sequencing of the human genome and is still relevant for targeted resequencing [27, 37, 61]. However, the advent of massively parallel sequencing (next-generation sequencing, NGS) turned out to be another game changer and revolutionized human genetics. Within 10 years, NGS led to a dramatic increase in knowledge on genetic variation and allowed fast and accurate diagnostics of clinically relevant germline and somatic mutations [45]. Different methods using semiconductors (Ion Torrent), pyrosequencing (Roche), sequencing by ligation (Applied Biosystems), and the widely used sequencing by synthesis with reversible terminators (Solexa, Illumina) enabled gene panel, whole-exome, and whole-genome sequencing within a few days at moderate costs [43]. However, both Sanger sequencing and NGS technologies deliver only short-read DNA fragments within the range of 50–1000 bases. The short-reads prevent analysis of complex genomic loci, repetitive elements, or variant phasing (haplotyping) and result in inefficient and incomplete genome assemblies. Moreover, PCR amplification of sequencing templates generates artefacts and precludes detection of native base modifications. Several of these shortcomings can be overcome by third-generation sequencing technologies (TGS), also referred to as long-read sequencing in the following.
Long-read next-generation sequencing methods
Nanopore sequencing
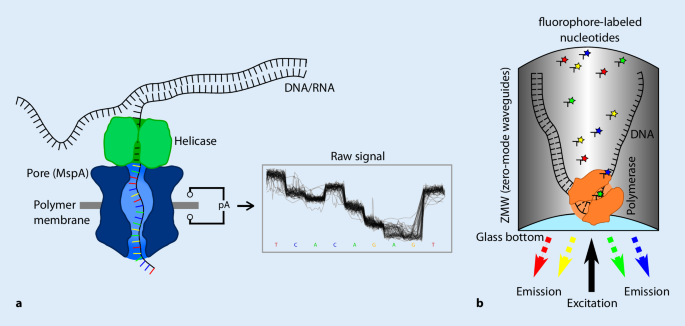
The idea to sequence long fragments of DNA and RNA without PCR amplification and nucleotide labeling had its origins as early as the 1980s, but has only become feasible after a technology using nanopores recently reached market maturity (Oxford Nanopore Technologies®, ONT, Oxford, UK) [14, 34]. In nanopore sequencing, a tiny protein pore (Mycobacterium smegmatis porin A, MspA, or Escherichia coli Curlin sigma S‑dependent growth subunit G, CsgG) is embedded in an electrically resistant polymer membrane and an ionic current is passed through this nanopore by setting a voltage across the membrane. When DNA or RNA passes through the pore via a helicase, this creates a characteristic change in the current, which provides information on the respective nucleotides in the nanopore (Fig. 1a; Table 1). The technology does not depend on a polymerase and allows sequencing of native DNA and RNA and the detection of various chemical modifications (e.g., methylation) of nucleic acids [12]. The longest reads achieved with the current method comprise a length of more than 2 million bases of DNA in a row.
Principle of nanopore and single-molecule real-time (SMRT) sequencing. a Nanopore sequencing: DNA is analyzed by threading it through a biological protein pore (e.g., Mycobacterium smegmatis porin A, MspA). The DNA is unzipped by a helicase to allow single-strand sequencing. Nucleotides inside the pore disrupt the ion flow through the channel. Each flow cell operates up to 50 (Flongle), 512 (MinION/GridION) or 3000 (PromethION) pores in parallel. DNA and RNA can be sequenced and processed in real-time at a speed of 450 (or 70 for RNA) bases per second and pore. The resulting current traces are converted to DNA sequences. b SMRT sequencing: The polymerase is attached to the bottom of a zero-mode waveguide (ZMW) µ‑well and incorporates fluorophore-labeled nucleotides. 100,000 (RSII), 1 million (Sequel) or 8 million (Sequel 2) of these µ‑wells are combined on one flow cell. During elongation with few bases per second, the fluorophore-labeled nucleotides are excited through a laser and emitted light is detected by four complementary metal oxide semiconductor cameras, one per color. The time-resolved fluorescence signals are converted to DNA sequences
SMRT sequencing
In single-molecule real-time (SMRT) sequencing, a single DNA polymerase molecule is immobilized at the bottom of picoliter wells called zero-mode waveguides (ZMWs). These wells are small enough to allow real-time recording of individual fluorescence signals on excitation by a laser when labeled nucleotides are progressively incorporated by the polymerase during the replication process (Fig. 1b; Table 1; [54]). The technology, commercialized by Pacific Biosciences® (Pacific Biosciences of California, Inc., Menlo Park, CA, USA), produces an average read length of 10–30 kb, but reads can exceed 80 kb [60]. Circular DNAs serve as a sequencing template and can be sequenced multiple times to provide higher accuracy consensus sequences. Base modifications affect the speed of nucleotide incorporation, which enables SMRT sequencing to detect modified bases.
Other approaches
Currently there are only a few alternatives to assessing long stretches of nucleic acids. Synthetic long read (SLR) technologies are offered by Illumina® or by emulsion-based sequencing from 10X Genomics®. However, both techniques are built on classical Illumina short-read sequencing and are in fact not TGS technologies. BioNano Genomics® uses an optical mapping method to mark sequences in long DNA fragments (500 bases – megabases) which are imaged and allow long-range genome mapping and detection of structural variants (Saphyr system).
Applications of long-read sequencing in human genetics
The first applications of long-read sequencing were restricted to the sequencing of smaller genomes such as bacteria. However, with improvements in chemistry, human genome sequencing became feasible [29]. In contrast to short-reads, these technologies enable unambiguous mapping of reads such as in regions of high homology, low complexity, or in pseudogenes. Also, the phasing of alleles (generation of haplotypes) is facilitated by long reads and is possible without information on the parental SNPs. This also allows whether genetic variants occur on the same allele or on opposite strands to be distinguished. Recent examples demonstrated that complete haplotyping of highly complex regions, including killer cell immunoglobin-like receptor (KIR) and human leukocyte antigen (HLA) loci can be performed using long-read technologies [1]. With improvements in the read lengths, as yet unresolved regions of the human genome, such as low-copy repeats, telomeres or centromeres (for sequencing of the Y‑chromosome centromere see [30]), become accessible [39].
An obvious advantage of long-read sequencing is the detection of structural variations (SVs), including the detection of balanced chromosomal rearrangements. There are several studies demonstrating the successful identification of constitutive [50], complex “chromothrypsis” [11], or somatic genomic rearrangements [16, 25]. Exact characterization of breakpoints for larger indels [36] or the detection of fusion gene products [32] are possible with long-read approaches. Long-read whole genome sequencing can identify thousands of SVs that may escape NGS and allows otherwise missed disease-causative genomic aberrations to be discovered [8, 12, 53]. The identification of SVs from TGS data may also require lower coverage than with NGS [11].
Long-read sequencing also enables studying larger repeat-expansions that escape PCR-based approaches. Repetitive elements can be evaluated with high precision, for example, for the FMR1-associated Fragile X‑syndrome repeat and determination of its repeat-stability-relevant AGG interruptions [3]. Larger repeats such as the facioscapulohumeral muscular dystrophy (FSHD)-associated D4Z4 repeat array have also been fully sequenced by TGS [44]. Using long-read sequencing, novel expansions of intronic TTTCA and TTTTA repeats of SAMD12 have been reported in benign adult familial myoclonic epilepsy [28] and repeat expansions in NOTCH2NLC have recently been associated with a neuronal intranuclear inclusion disease [57]. The highly similar sequences of the tandem repeats can be directly assessed from the raw signal (Fig. 2). Cas9-based enrichments, e.g., of disease-causing repetitive or other genomic regions make TGS more feasible for routine diagnostic applications and allow several genomic loci to be analyzed in one assay. Utilizing the ONT Flongle for these targeted approaches enables the costs of TGS-based analysis to be further reduced.
Detection of tandem repeat expansions from nanopore sequencing raw signal traces. a, d Sample plots showing the raw nanopore sequencing signal from a tandem repeat expansion. a positive and d negative strands. The repeat consists of two distinct sequence motives, which are indicated by red and blue. Adjacent sequences are shown in gray. b, c, e, f Current profile of a single repeat unit. Source: Institute of Human Genetics, RWTH Aachen
The feasibility of long-read sequencing to detect unusual mutation mechanisms was recently reported for the exonization of an intronic LINE-1 element inserted into the DMD gene in a patient with muscular dystrophy [24]. Another example of an unusual mutation is a SINE-VNTR-Alu (SVA) retrotransposition into intron 32 of the TAF1 locus, which causes an endemic type of X‑linked dystonia parkinsonism [2].
Previous sequencing technologies provided only limited access to the state of nucleic acid modifications. In principle, any base modification that affects the current in nanopore sequencing (Fig. 3) or the nucleotide incorporation time in SMRT sequencing is recorded in the raw signals. It allows, for example, discrimination between 5‑methylcytosine and 5‑hydroxymethylcytosine, or detection of N6-methyladenosine [48, 56]. This unique feature of TGS enables SV, SNV, and the methylation status of genomic loci to be analyzed in parallel and may improve the molecular diagnostics, for example, of cancer and imprinting disorders. Not only the landscape of alternative splicing can be investigated by reading through entire isoforms [33], but the various base modifications present on native RNA molecules can also be detected using this PCR-free method [18]. Moreover, native CpG methylation and chromatin accessibility can be studied in parallel using long reads [38]. Table 2 provides an overview of current long-read sequencing applications.
Detection of native epigenetic modifications by nanopore sequencing. Methylation of a cytosine (C) causes a change in the recorded current profile. The signal of the unmodified cytosine is marked by a blue box and 5‑methylcytosine is labelled by a red box. Source: Institute of Human Genetics, RWTH Aachen
Challenges of long-read sequencing
Preparing of libraries for long-read sequencing is straightforward; however, there are several pitfalls in terms of obtaining optimal sequencing libraries. A major drawback of SMRT sequencing is the fixed number of µ‑wells per flow cell, which means that shorter or no sequencing templates per well reduce the overall output. In contrast, individual pores in nanopore sequencing can sequence up to several thousand molecules; however, very large DNA molecules tend to block respective pores. A major challenge in TGS sequencing is the high sequencing error rate, but higher coverage and optimized filtering strategies can improve consensus accuracy [14]. The release of a new ONT “linear consensus sequencing” (LCS) chemistry will provide better results, such as the “circular consensus sequencing” (CCS) chemistry used by PacBio. Another issue is the relatively large raw data file size, which requires a high demand for data management and storage especially for nanopore sequencing applications. PCR-free target enrichment strategies for nanopore sequencing are hardly available, but interesting approaches using CRISPR/Cas9 are under development. Cas9 is used to cleave and directly capture genomic regions via hybridization and immobilization on beads before sequencing. Moreover, software applications for nanopore sequencing may be useful for in silico target enrichment. ‘ReadUntil’ is a software application that allows fragments of interest to be selected by reversing the voltage across utilized nanopores and extruding DNA on the fly [41]. Bioinformatics strategies for the processing of long-read sequencing data are rapidly evolving; however, it is currently unclear which applications are the most suitable [52]. Notably, base calling performance is lower for modified bases owing to the lack of suited reference sequences and computational models. Table 3 provides an overview of some of the most commonly used bioinformatics tools in long-read sequencing.
Outlook
Long-read sequencing has a huge potential and will provide additional insight into genome biology and human genetics. Several disease-relevant genes and pathomechanisms that escape short-read sequencing technologies will be elucidated by long-read technologies. The technologies will soon become an integral part of molecular genetic diagnostics. An open question is whether the techniques will mature such that they will even replace short-read sequencing technologies, array-based analyses, and cytogenetics. Applications of TGS to detect SVs and tandem repeats are already superior to NGS and almost ready for use in molecular routine diagnostics. In contrast, the higher error rate of nanopore sequencing currently makes SNV detection only suitable in targeted sequencing approaches that generate a high coverage (> 100×). The lack of commercially available kits for TGS enrichments and gold-standard bioinformatics solutions is at the moment one of the bottlenecks for usage in molecular diagnostics. Besides the aforementioned applications, the portability of small nanopore sequencers opens up additional opportunities for field applications in a nearly lab-free environment. This is illustrated by surveillance of pathogens in disease epidemics, such as the real-time tracking of Ebola distribution [47] or the molecular mapping of Zika virus spread in Brazil [17]. Are we perhaps heading for times of “sequencing at home” or in outpatient clinics and medical practices, with direct data transfer to genetic specialists? Other open questions concern the speed of nanopore technologies from library preparation to obtaining the first sequencing results within minutes to a few hours: Can we tackle fast sepsis diagnostics or intraoperative molecular genotyping? Undoubtedly, genetics is becoming increasingly important in many fields of health care and the possibilities for addressing the plentiful questions by TGS are rapidly evolving.
Conclusions for clinical practice
-
Different long-read sequencing platforms are available that either depend on an immobilized polymerase and fluorescently labelled nucleotides or on biological (nano)pores.
-
Long-read sequencing is mostly applied in research, but has the potential to be used in many fields of molecular genetic diagnostics.
-
Long-read sequencing has several advantages compared with short-read sequencing methods and is well suited to, for example, addressing structural variations, epigenetic modifications, and repetitive elements of the genome.
References
Ameur A, Kloosterman WP, Hestand MS (2019) Single-molecule sequencing: towards clinical applications. Trends Biotechnol 37:72–85
Aneichyk T, Hendriks WT, Yadav R et al (2018) Dissecting the causal mechanism of X‑linked Dystonia-parkinsonism by integrating genome and transcriptome assembly. Cell 172:897–909e21
Ardui S, Race V, Zablotskaya A et al (2017) Detecting AGG interruptions in male and female FMR1 premutation carriers by single-molecule sequencing. Hum Mutat 38:324–331
Ardui S, Ameur A, Vermeesch JR, Hestand MS (2018) Single molecule real-time (SMRT) sequencing comes of age: applications and utilities for medical diagnostics. Nucleic Acids Research 46(5):2159–2168. https://doi.org/10.1093/nar/gky066
Borràs DM, Vossen RHAM, Liem M, Buermans HPJ, Dauwerse H, van Heusden D, Gansevoort RT, den JT Dunnen, Janssen B, Peters DJM, Losekoot M, Anvar SY (2017) Detecting variants in polycystic kidney disease patients by single-molecule long-read sequencing. Human Mutation 38(7):870–879
Brønstad Brynildsrud O, Eldholm V, Bohlin J, Uadiale K, Obaro S, Caugant SA (2018) Acquisition of virulence genes by a carrier strain gave rise to the ongoing epidemics of meningococcal disease in West Africa. Proceedings of the National Academy of Sciences 115(21):5510–5515
Břinda K, Hanage WP et al (2018) Lineage calling can identify antibiotic resistant clones within minutes. bioRxiv 403204
Chaisson MJP, Sanders AD, Zhao X et al (2019) Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat Commun 10:1784
Clark MB, Tunbridge EM et al (2018) Long-read sequencing reveals the splicing profile of the calcium channel gene CACNA1C in human brain. bioRxiv 260562
Cornelis S, Gansemans Y, Vander Plaetsen A‑S, Weymaere J, Willems S, Deforce D, Van Nieuwerburgh F (2019) Forensic tri-allelic SNP genotyping using nanopore sequencing. Forensic Science International: Genetics 38:204–210
Cretu Stancu M, Van Roosmalen MJ, Renkens I et al (2017) Mapping and phasing of structural variation in patient genomes using nanopore sequencing. Nat Commun 8:1326
De Coster W, De Roeck A, De Pooter T et al (2018) Structural variants identified by Oxford Nanopore PromethION sequencing of the human genome. bioRxiv:434118
De Roeck A, Duchateau L, Van Dongen J, Cacace R, Bjerke M, Van den Bossche T, Cras P, Vandenberghe R, De Deyn PP, Engelborghs S, Van Broeckhoven C, Sleegers K (2018) An intronic VNTR affects splicing of ABCA7 and increases risk of Alzheimer’s disease. Acta Neuropathologica 135(6):827–837
Deamer D, Akeson M, Branton D (2016) Three decades of nanopore sequencing. Nat Biotechnol 34:518–524
Dutta UR, Rao SN, Pidugu VK, Vineeth VS, Bhattacherjee A, Bhowmik AD, Ramaswamy SK, Singh KG, Dalal A (2018) Breakpoint mapping of a novel de novo translocation t(X;20)(q11.1;p13) by positional cloning and long read sequencing. Genomics. https://doi.org/10.1016/j.ygeno.2018.07.005
Euskirchen P, Bielle F, Labreche K et al (2017) Same-day genomic and epigenomic diagnosis of brain tumors using real-time nanopore sequencing. Acta Neuropathol 134:691–703
Faria NR, Quick J, Claro IM et al (2017) Establishment and cryptic transmission of Zika virus in Brazil and the Americas. Nature 546:406–410
Garalde DR, Snell EA, Jachimowicz D et al (2018) Highly parallel direct RNA sequencing on an array of nanopores. Nat Methods 15:201–206
George S, Dingle KE et al (2018) MinION nanopore sequencing of multiple displacement amplified mycobacteria DNA direct from sputum. bioRxiv 490417
Gigante S, Ritchie ME et al (2018) Using long-read sequencing to detect imprinted DNA methylation. bioRxiv 445924
Gilbert W, Maxam A (1973) The nucleotide sequence of the lac operator. Proc Natl Acad Sci U S A 70:3581–3584
Gilpatrick T, Timp W et al (2019) Targeted nanopore sequencing with Cas9 for studies of methylation, structural variants and mutations. bioRxiv 604173
Golparian D, Donà V, Sánchez-Busó L, Foerster S, Harris S, Endimiani A, Low N, Unemo M (2018) Antimicrobial resistance prediction and phylogenetic analysis of Neisseria gonorrhoeae isolates using the Oxford Nanopore MinION sequencer. Scientific Reports. https://doi.org/10.1038/s41598-018-35750-4
Goncalves A, Oliveira J, Coelho T et al (2017) Exonization of an Intronic LINE-1 element causing Becker muscular dystrophy as a novel mutational mechanism in dystrophin gene. Genes (Basel). https://doi.org/10.3390/genes8100253
Gong L, Wong CH, Cheng WC et al (2018) Picky comprehensively detects high-resolution structural variants in nanopore long reads. Nat Methods 15:455–460
Grubaugh ND, Gangavarapu K, Quick J, Matteson NL, Goes De Jesus J, Main BJ, Tan AL, Paul LM, Brackney DE, Grewal S, Gurfield N, van Rompay KKA, Isern S, Michael SF, Coffey LL, Loman NJ, Andersen KG (2019) An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biology. https://doi.org/10.1186/s13059-018-1618-7
International Human Genome Sequencing C (2004) Finishing the euchromatic sequence of the human genome. Nature 431:931–945
Ishiura H, Doi K, Mitsui J et al (2018) Expansions of intronic TTTCA and TTTTA repeats in benign adult familial myoclonic epilepsy. Nat Genet 50:581–590
Jain M, Koren S, Miga KH et al (2018a) Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat Biotechnol 36:338–345
Jain M, Olsen HE, Turner DJ et al (2018b) Linear assembly of a human centromere on the Y chromosome. Nat Biotechnol 36:321–323
Jeck WR, Lee J, Robinson H, Le LP, Iafrate AJ, Nardi V (2019) A nanopore sequencing–based assay for rapid detection of gene fusions. J Molecul Diagn 21(1):58–69
Jeck WR, Lee J, Robinson H et al (2019) A Nanopore sequencing-based assay for rapid detection of gene fusions. J Mol Diagn 21:58–69
Karsai G, Kraft F, Haag N et al (2019) DEGS1-associated aberrant sphingolipid metabolism impairs nervous system function in humans. J Clin Invest 129:1229–1239
Kasianowicz JJ, Bezrukov SM (2016) On ‘three decades of nanopore sequencing’. Nat Biotechnol 34:481–482
Kerkhof LJ, Dillon KP, Häggblom MM, McGuinness LR (2017) Profiling bacterial communities by MinION sequencing of ribosomal operons. Microbiome. https://doi.org/10.1186/s40168-017-0336-9
Kraft F, Wesseler K, Begemann M et al (2019) Novel familial distal imprinting centre 1 (11p15.5) deletion provides further insights in imprinting regulation. Clin Epigenetics 11:30
Lander ES, Linton LM, Birren B et al (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921
Lee I, Razaghi R, Gilpatrick T et al (2018) Simultaneous profiling of chromatin accessibility and methylation on human cell lines with nanopore sequencing. bioRxiv:504993
Li W, Freudenberg J (2014) Mappability and read length. Front Genet 5:381
Liau Y, Cree SL et al (2019) Nanopore sequencing of the pharmacogene CYP2D6 allows simultaneous haplotyping and detection of duplications. bioRxiv 576280
Loose M, Malla S, Stout M (2016) Real-time selective sequencing using nanopore technology. Nat Methods 13:751–754
Maxam AM, Gilbert W (1977) A new method for sequencing DNA. Proc Natl Acad Sci U S A 74:560–564
Metzker ML (2010) Sequencing technologies—the next generation. Nat Rev Genet 11:31–46
Mitsuhashi S, Nakagawa S, Takahashi Ueda M et al (2017) Nanopore-based single molecule sequencing of the D4Z4 array responsible for facioscapulohumeral muscular dystrophy. Sci Rep 7:14789
Ng SB, Buckingham KJ, Lee C et al (2010) Exome sequencing identifies the cause of a mendelian disorder. Nat Genet 42:30–35
Nicholls SM, Quick JC, Tang S, Loman NJ (2019) Ultra-deep, long-read nanopore sequencing of mock microbial community standards. GigaScience. https://doi.org/10.1093/gigascience/giz043
Quick J, Loman NJ, Duraffour S et al (2016) Real-time, portable genome sequencing for Ebola surveillance. Nature 530:228–232
Rand A, Jain M, Eizenga J et al (2017) Mapping DNA methylation with high-throughput nanopore sequencing. Nat Methods 14:411–413
Roe D, Vierra-Green C, Pyo C‑W, Eng K, Hall R, Kuang R, Spellman S, Ranade S, Geraghty DE, Maiers M (2017) Revealing complete complex KIR haplotypes phased by long-read sequencing technology. Genes & Immunity 18(3):127–134
Sanchis-Juan A, Stephens J, French CE et al (2018) Complex structural variants in Mendelian disorders: identification and breakpoint resolution using short- and long-read genome sequencing. Genome Med 10:95
Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A 74:5463–5467
Sedlazeck FJ, Lee H, Darby CA et al (2018) Piercing the dark matter: bioinformatics of long-range sequencing and mapping. Nat Rev Genet 19:329–346
Seo JS, Rhie A, Kim J et al (2016) De novo assembly and phasing of a Korean human genome. Nature 538:243–247
Shendure J, Balasubramanian S, Church GM et al (2017) DNA sequencing at 40: past, present and future. Nature 550:345–353
Shin J, Lee S, Go M‑J, Lee SY, Kim SC, Lee C‑H, Cho B‑K (2016) Analysis of the mouse gut microbiome using full-length 16S rRNA amplicon sequencing. Scientific Reports. https://doi.org/10.1038/srep29681
Simpson JT, Workman RE, Zuzarte PC et al (2017) Detecting DNA cytosine methylation using nanopore sequencing. Nat Methods 14:407–410
Sone J, Mitsuhashi S, Fujita A et al (2019) Long-read sequencing identifies GGC repeat expansion in human-specific NOTCH2NLC associated with neuronal intranuclear inclusion disease. bioRxiv:515635
Tang AD, Brooks AN et al (2018) Full-length transcript characterization of SF3B1 mutation in chronic lymphocytic leukemia reveals downregulation of retained introns. bioRxiv 410183
Ton KNT, Cree SL, Gronert-Sum SJ, Merriman TR, Stamp LK, Kennedy MA (2018) Multiplexed nanopore sequencing of HLA-B locus in Māori and Pacific island samples. Frontiers in Genetics. https://doi.org/10.3389/fgene.2018.00152
Van Dijk EL, Jaszczyszyn Y, Naquin D et al (2018) The third revolution in sequencing technology. Trends Genet 34:666–681
Venter JC, Adams MD, Myers EW et al (2001) The sequence of the human genome. Science 291:1304–1351
Wu R, Kaiser AD (1968) Structure and base sequence in the cohesive ends of bacteriophage lambda DNA. J Mol Biol 35:523–537
Acknowledgements
We apologize that many outstanding papers in the field have not been cited owing to the limitations of space. We would like to point out that developments in the field of long-read sequencing illustrate changes in the current research practice toward rapid publication of results on preprint servers such as bioRXiv (https://www.biorxiv.org/), the nanopore community platform (https://nanoporetech.com/community), Twitter, or as blogs. In our opinion, this practice fosters lively discussion and speedy innovation, and may serve as a contemporary model to complement the often viscous and delaying peer-review processes.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
F. Kraft and I. Kurth declare that they have no competing interests.
For this article no studies with human participants or animals were performed by any of the authors. All studies performed were in accordance with the ethical standards indicated in each case.
Rights and permissions
Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Kraft, F., Kurth, I. Long-read sequencing in human genetics. medgen 31, 198–204 (2019). https://doi.org/10.1007/s11825-019-0249-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11825-019-0249-z
Keywords
- Third-generation sequencing
- Long-read sequencing
- Nanopore sequencing
- Single-molecule real-time sequencing
- Genomics