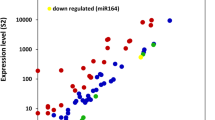
Abstract
The miR396 and growth-regulating factor (GRF) regulatory network is evolutionarily conserved among plant species. The orthologous GRF genes in rice were involved in regulating grain development, and targeted by miR396. However, there are still no more information about the involvement of miR396 and GRF genes in maize grain. In this article, RNA sequencing was performed on the developing maize kernels to analyze the dynamic profiles of miR396 and GRF genes and explore their possible regulatory roles during maize effective grain filling. Our results showed that GRF genes were also the conserved targets of miR396 in maize, according to computational prediction and validated by degradome sequencing. MiR396 expressed high and gradually declined with advancing maize grain filling, significant negatively correlated with its target GRF genes, which mostly increased continuously. Our test also provided a testimony of miR396-GRF network in different regulations between the development of maize embryo and endosperm by real-time quantitative PCR. And further analysis of expression pattern suggested that miR319, miR166, and RDR might interact with miR396-GRF network during grain development in maize. This study mainly provided a valuable foundation for future comprehensive analysis of miR396-GRF network, and further research work is needed to confirm the regulatory roles of miR396 and GRF genes during grain filling in maize.



Similar content being viewed by others
References
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297
Bonnet E, He Y, Billiau K, Van de Peer Y (2010) TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics 26(12):1566–1568
Borrás L, Westgate ME (2006) Predicting maize kernel sink capacity early in development. Field Crop Res 95(2–3):223–233
CF J, AJ R (1972) Factors limiting the supply of sucrose to the developing wheat grain. Ann Bot 36:729–741
Choi D, Kim JH, Kende H (2004) Whole genome analysis of the OsGRF gene family encoding plant-specific putative transcription activators in rice (Oryza sativa L.). Plant Cell Physiol 45(7):897–904
Cuperus JT, Fahlgren N, Carrington JC (2011) Evolution and functional diversification of MIRNA genes. Plant Cell 23(2):431–442
Daszkowska-Golec A, Szarejko I (2013) The molecular basis of ABA-mediated plant response to drought
De Smet I, Lau S, Mayer U, Jurgens G (2010) Embryogenesis–the humble beginnings of plant life. Plant J 61(6):959–970
Debernardi JM, Rodriguez RE, Mecchia MA, Palatnik JF (2012) Functional specialization of the plant miR396 regulatory network through distinct microRNA-target interactions. PLoS Genet 8(1):e1002419
Ding Y, Chen Z, Zhu C (2011) Microarray-based analysis of cadmium-responsive microRNAs in rice (Oryza sativa). J Exp Bot 62(10):3563–3573
Ding D, Wang Y, Han M, Fu Z, Li W, Liu Z, Hu Y, Tang J (2012) MicroRNA transcriptomic analysis of heterosis during maize seed germination. PLoS One 7(6):e39578
DR M (1995) Genetic control and integration of maturation and germination pathways in seed development. Ann Rev Plant Physiol Plant Mol Biol 46:71–93
Fang R, Li L, Li J (2013) Spatial and temporal expression modes of MicroRNAs in an elite rice hybrid and its parental lines. Planta 238(2):259–269
Galla G, Volpato M, Sharbel TF, Barcaccia G (2013) Computational identification of conserved microRNAs and their putative targets in the Hypericum perforatum L. flower transcriptome. Plant Reprod 26(3):209–229
Gore MA, Chia JM, Elshire RJ, Sun Q, Ersoz ES, Hurwitz BL, Peiffer JA, McMullen MD, Grills GS, Ross-Ibarra J, Ware DH, Buckler ES (2009) A first-generation haplotype map of maize. Science 326(5956):1115–1117
Gu Y, Liu Y, Zhang J, Liu H, Hu Y, Du H, Li Y, Chen J, Wei B, Huang Y (2013) Identification and characterization of microRNAs in the developing maize endosperm. Genomics 102(5–6):472–478
Hammond SM, Bernstein E, Beach D, Hannon GJ (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404:293–296
Horiguchi G, Kim GT, Tsukaya H (2005) The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J 43(1):68–78
Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14(6):787–799
Kim JH, Kende H (2004) A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc Natl Acad Sci USA 101(36):13374–13379
Kim JH, Choi D, Kende H (2003) The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J 36(1):94–104
Kruszka K, Pieczynski M, Windels D, Bielewicz D, Jarmolowski A, Szweykowska-Kulinska Z, Vazquez F (2012) Role of microRNAs and other sRNAs of plants in their changing environments. J Plant Physiol 169(16):1664–1672
Lan Y, Su N, Shen Y, Zhang R, Wu F, Wan J (2012) Identification of novel MiRNAs and MiRNA expression profiling during grain development in indica rice. BMC Genom 13:264
Liu D, Song Y, Chen Z, Yu D (2009) Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol Plant 136(2):223–236
Liu ZH, Ji HQ, Cui ZT, Wu X, Duan LJ, Feng XX, Tang JH (2010) QTL detected for grain-filling rate in maize using a RIL population. Mol Breed 27(1):25–36
Luo M, Gao J, Peng H, Pan G, Zhang Z (2014) MiR393-targeted TIR1-like (F-box) gene in response to inoculation to R. Solani in Zea mays. Acta Physiologiae Plantarum 36(5):1283–1291
Lur H-S, Setter T (1993) Role of Auxin in Maize Endosperm Development. Plant Physiol 103:273–280
Martin-Trillo M, Cubas P (2010) TCP genes: a family snapshot ten years later. Trends Plant Sci 15:31–39
Mecchia MA, Debernardi JM, Rodriguez RE, Schommer C, Palatnik JF (2013) MicroRNA miR396 and RDR6 synergistically regulate leaf development. Mech Dev 130(1):2–13
Mechin V, Balliau T, Chateau-Joubert S, Davanture M, Langella O, Negroni L, Prioul JL, Thevenot C, Zivy M, Damerval C (2004) A two-dimensional proteome map of maize endosperm. Phytochemistry 65(11):1609–1618
Peng T, Sun H, Du Y, Zhang J, Li J, Liu Y, Zhao Y, Zhao Q (2013) Characterization and expression patterns of microRNAs involved in rice grain filling. PLoS One 8(1):e54148
Prioul JL, Mechin V, Lessard P, Thevenot C, Grimmer M, Chateau-Joubert S, Coates S, Hartings H, Kloiber-Maitz M, Murigneux A, Sarda X, Damerval C, Edwards KJ (2008) A joint transcriptomic, proteomic and metabolic analysis of maize endosperm development and starch filling. Plant Biotechnol J 6(9):855–869
Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002) Prediction of plant microRNA targets. Cell 110(4):513–520
Rodriguez RE, Mecchia MA, Debernardi JM, Schommer C, Weigel D, Palatnik JF (2010) Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137(1):103–112
Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326(5956):1112–1115
Sunkar R, Jagadeeswaran G (2008) In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biol 8:37
Takai T, Fukuta Y, Shiraiwa T, Horie T (2005) Time-related mapping of quantitative trait loci controlling grain-filling in rice (Oryza sativa L.). J Exp Bot 56(418):2107–2118
Teoh KT, Requesens DV, Devaiah SP, Johnson D, Huang X, Howard JA, Hood EE (2013) Transcriptome analysis of embryo maturation in maize. BMC Plant Biol 13:19
van der Knaap E, Kim JH, Kende H (2000) A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol 122(3):695–704
Voinnet O (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136(4):669–687
Wang L, Gu X, Xu D, Wang W, Wang H, Zeng M, Chang Z, Huang H, Cui X (2011) miR396-targeted AtGRF transcription factors are required for coordination of cell division and differentiation during leaf development in Arabidopsis. J Exp Bot 62(2):761–773
Ye R, Yao Q, Xu Z, Xue H (2004) Development of an efficient method for the isolation of factors involved in gene transcription during rice embryo development. Plant J 38:348–357
Yi R, Zhu Z, Hu J, Qian Q, Dai J, Ding Y (2013) Identification and expression analysis of microRNAs at the grain filling stage in rice(Oryza sativa L.)via deep sequencing. PLoS One 8(3):e57863
Zhang D-F, Li B, Jia G-Q, Zhang T-F, Dai J-R, Li J-S, Wang S-C (2008) Isolation and characterization of genes encoding GRF transcription factors and GIF transcriptional coactivators in Maize (Zea mays L.). Plant Sci 175(6):809–817
Zhang W, Sun P, He Q, Shu F, Wang J, Deng H (2013) Fine mapping of GS2, a dominant gene for big grain rice. Crop J 1(2):160–165
Acknowledgments
This work was supported by a grant from the National Nature Foundation of China (31370033).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E. Schleiff.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11738_2014_1767_MOESM1_ESM.tif
Supplementary material 1 Fig. S1 Representation of the GO-term analysis for miR396 targeted GRF genes. The most notable GO categories are ATP binding, nucleus, hydrolase activity and regulation of transcription. (TIFF 24 kb)
11738_2014_1767_MOESM2_ESM.tif
Supplementary material 2 Fig. S2 Representation of purine pathway (map00230) in which GIF2 involved. The red rectangle representing RNA polymerase (EC: 2. 7. 7. 6) of purine pathway coded by GIF2 gene. (TIFF 115 kb)
11738_2014_1767_MOESM3_ESM.tif
Supplementary material 3 Fig. S3 Representation of pyrimidine pathway (map00240) in which GIF2 involved. The red rectangle representing RNA polymerase (EC: 2. 7. 7. 6) of pyrimidine pathway coded by GIF2 gene. (TIFF 80 kb)
11738_2014_1767_MOESM4_ESM.doc
Supplementary material 3 Table S1 Summary of small RNAs classes during maize grain filling. Table S2 The targets of miR396, miR166 and miR319 identified by degradome sequencing in this study. Table S3 The different expression of miRNA (RPM) during maize kernel development from 15 to 35 DAP. Table S4 The different expression of GIFs and GRFs (RPKM) during maize kernel development from 15 to 35 DAP. A: the different expression of GIFs and GRFs in maize line Chang7-2. B: the different expression of GIFs and GRFs in maize line Zheng58. Table S5 The different expressed target genes of miRNA during maize kernel development from 15 to 35 DAP, which might interact with miR396. (DOC 208 kb)
Rights and permissions
About this article
Cite this article
Zhang, K., Shi, X., Zhao, X. et al. Investigation of miR396 and growth-regulating factor regulatory network in maize grain filling. Acta Physiol Plant 37, 28 (2015). https://doi.org/10.1007/s11738-014-1767-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-014-1767-6