Abstract
Background
Genetic factors alter the risk for nonalcoholic fatty liver disease (NAFLD). We sought to identify NAFLD-associated genes and elucidate gene networks and pathways involved in the pathogenesis of NAFLD.
Methods
Quantitative global hepatic gene expression analysis was performed on 53 morbidly obese Caucasian subjects undergoing bariatric surgery (27 with NAFLD and 26 controls). After standardization of data, gene expression profiles were compared between patients with NAFLD and controls. The set of genes that significantly correlated with NAFLD was further analyzed by hierarchical clustering and ingenuity pathways analyses.
Results
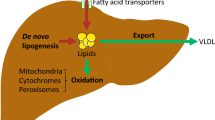
There were 25,643 quantitative transcripts, of which 108 were significantly associated with NAFLD (p < 0.001). Canonical pathway analysis in the NAFLD-associated gene clusters showed that the hepatic fibrosis signaling was the most significant pathway in the up-regulated NAFLD gene cluster containing three (COL1A1, IL10, IGFBP3) significantly altered genes, whereas the endoplasmic reticulum stress and protein ubiquitination pathways were the most significant pathways in the down-regulated NAFLD gene cluster, with the first pathway containing one (HSPA5) and the second containing two (HSPA5, USP25) significantly altered genes. The four primary gene networks associated with NAFLD were involved in cell death, immunological disease, cellular movement, and lipid metabolism with several significantly altered “hub” genes in these networks.
Conclusions
This study reveals the canonical pathways and gene networks associated with NAFLD in morbidly obese Caucasians. The application of gene network analysis highlights the transcriptional relationships among NAFLD-associated genes and allows identification of hub genes that may represent high-priority candidates for NAFLD.



Similar content being viewed by others
References
Schwimmer JB, Deutsch R, Kahen T, et al. Prevalence of fatty liver in children and adolescents. Pediatrics. 2006;118(4):1388–93.
Clark JM, Brancati FL, Diehl AM. The prevalence and etiology of elevated aminotransferase levels in the United States. Am J Gastroenterol. 2003;98(5):960–7.
Charlton M. Nonalcoholic fatty liver disease: a review of current understanding and future impact. Clin Gastroenterol Hepatol. 2004;2(12):1048–58.
Sanyal AJ. AGA technical review on nonalcoholic fatty liver disease. Gastroenterology. 2002;123(5):1705–25.
Brunt EM. Nonalcoholic steatohepatitis. Semin Liver Dis. 2004;24(1):3–20.
Matteoni CA, Younossi ZM, Gramlich T, et al. Nonalcoholic fatty liver disease: a spectrum of clinical and pathological severity. Gastroenterology. 1999;116(6):1413–9.
Ekstedt M, Franzen LE, Mathiesen UL, et al. Long-term follow-up of patients with NAFLD and elevated liver enzymes. Hepatology. 2006;44(4):865–73.
Lazo M, Clark JM. The epidemiology of nonalcoholic fatty liver disease: a global perspective. Semin Liver Dis. 2008;28(4):339–50.
Browning JD, Horton JD. Molecular mediators of hepatic steatosis and liver injury. J Clin Invest. 2004;114(2):147–52.
Dixon JB, Bhathal PS, O'Brien PE. Nonalcoholic fatty liver disease: predictors of nonalcoholic steatohepatitis and liver fibrosis in the severely obese. Gastroenterology. 2001;121(1):91–100.
Younossi ZM, Baranova A, Ziegler K, et al. A genomic and proteomic study of the spectrum of nonalcoholic fatty liver disease. Hepatology. 2005;42(3):665–74.
Struben VM, Hespenheide EE, Caldwell SH. Nonalcoholic steatohepatitis and cryptogenic cirrhosis within kindreds. Am J Med. 2000;108(1):9–13.
Willner IR, Waters B, Patil SR, et al. Ninety patients with nonalcoholic steatohepatitis: insulin resistance, familial tendency, and severity of disease. Am J Gastroenterol. 2001;96(10):2957–61.
Schwimmer JB, Celedon MA, Lavine JE, et al. Heritability of nonalcoholic fatty liver disease. Gastroenterology. 2009;136(5):1585–92.
Browning JD, Szczepaniak LS, Dobbins R, et al. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 2004;40(6):1387–95.
Browning JD, Kumar KS, Saboorian MH, et al. Ethnic differences in the prevalence of cryptogenic cirrhosis. Am J Gastroenterol. 2004;99(2):292–8.
Miele L, Beale G, Patman G, et al. The Kruppel-like factor 6 genotype is associated with fibrosis in nonalcoholic fatty liver disease. Gastroenterology. 2008;135(1):282–91.
Romeo S, Kozlitina J, Xing C, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40(12):1461–5.
Yoneda M, Hotta K, Nozaki Y, et al. Association between PPARGC1A polymorphisms and the occurrence of nonalcoholic fatty liver disease (NAFLD). BMC Gastroenterol. 2008;8:27.
Sreekumar R, Rosado B, Rasmussen D, et al. Hepatic gene expression in histologically progressive nonalcoholic steatohepatitis. Hepatology. 2003;38(1):244–51.
Chiappini F, Barrier A, Saffroy R, et al. Exploration of global gene expression in human liver steatosis by high-density oligonucleotide microarray. Lab Invest. 2006;86(2):154–65.
Yoneda M, Endo H, Mawatari H, et al. Gene expression profiling of non-alcoholic steatohepatitis using gene set enrichment analysis. Hepatol Res. 2008;38(12):1204–12.
Greco D, Kotronen A, Westerbacka J, et al. Gene expression in human NAFLD. Am J Physiol Gastrointest Liver Physiol. 2008;294(5):G1281–7.
Kleiner DE, Brunt EM, Van Natta M, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41(6):1313–21.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300.
Elam MB, Cowan Jr GS, Rooney RJ, et al. Hepatic gene expression in morbidly obese women: implications for disease susceptibility. Obesity (Silver Spring). 2009;17(8):1563–73.
Maiti AK. Gene network analysis of oxidative stress-mediated drug sensitivity in resistant ovarian carcinoma cells. Pharmacogenomics J. 2010;10(2):94–104.
Goring HH, Curran JE, Johnson MP, et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet. 2007;39(10):1208–16.
Eisen MB, Spellman PT, Brown PO, et al. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95(25):14863–8.
Furuhashi M, Hotamisligil GS. Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat Rev Drug Discov. 2008;7(6):489–503.
Furuhashi M, Fucho R, Gorgun CZ, et al. Adipocyte/Macrophage fatty acid-binding proteins contribute to metabolic deterioration through actions in both macrophages and adipocytes in mice. J Clin Invest. 2008;118(7):2640–50.
Tuncman G, Erbay E, Hom X, et al. A genetic variant at the fatty acid-binding protein aP2 locus reduces the risk for hypertriglyceridemia, type 2 diabetes, and cardiovascular disease. Proc Natl Acad Sci U S A. 2006;103(18):6970–5.
Maeda K, Cao H, Kono K, et al. Adipocyte/Macrophage fatty acid binding proteins control integrated metabolic responses in obesity and diabetes. Cell Metab. 2005;1(2):107–19.
Serra-Pages C, Medley QG, Tang M, et al. Liprins, a family of LAR transmembrane protein-tyrosine phosphatase-interacting proteins. J Biol Chem. 1998;273(25):15611–20.
Kriajevska M, Fischer-Larsen M, Moertz E, et al. Liprin beta 1, a member of the family of LAR transmembrane tyrosine phosphatase-interacting proteins, is a new target for the metastasis-associated protein S100A4 (Mts1). J Biol Chem. 2002;277(7):5229–35.
Yang JJ. Mixed lineage kinase ZAK utilizing MKK7 and not MKK4 to activate the c-Jun N-terminal kinase and playing a role in the cell arrest. Biochem Biophys Res Commun. 2002;297(1):105–10.
Jandhyala DM, Ahluwalia A, Obrig T, et al. ZAK: a MAP3Kinase that transduces Shiga toxin- and ricin-induced proinflammatory cytokine expression. Cell Microbiol. 2008;10(7):1468–77.
Cheng YC, Kuo WW, Wu HC, et al. ZAK induces MMP-2 activity via JNK/p38 signals and reduces MMP-9 activity by increasing TIMP-1/2 expression in H9c2 cardiomyoblast cells. Mol Cell Biochem. 2009;325(1–2):69–77.
Friedman SL. Molecular regulation of hepatic fibrosis, an integrated cellular response to tissue injury. J Biol Chem. 2000;275(4):2247–50.
Yamaguchi M. Role of calcium-binding protein regucalcin in regenerating rat liver. J Gastroenterol Hepatol. 1998;13(Suppl):S106–12.
Izumi T, Yamaguchi M. Overexpression of regucalcin suppresses cell death and apoptosis in cloned rat hepatoma H4-II-E cells induced by lipopolysaccharide, PD 98059, dibucaine, or Bay K 8644. J Cell Biochem. 2004;93(3):598–608.
Fukaya Y, Yamaguchi M. Overexpression of regucalcin suppresses cell death and apoptosis in cloned rat hepatoma H4-II-E cells induced by insulin or insulin-like growth factor-I. J Cell Biochem. 2005;96(1):145–54.
Matsubara M, Tanaka T, Terato H, et al. Action mechanism of human SMUG1 uracil-DNA glycosylase. Nucleic Acids Symp Ser (Oxf). 2005;49(49):295–6.
Pettersen HS, Sundheim O, Gilljam KM, et al. Uracil-DNA glycosylases SMUG1 and UNG2 coordinate the initial steps of base excision repair by distinct mechanisms. Nucleic Acids Res. 2007;35(12):3879–92.
Seki S, Kitada T, Yamada T, et al. In situ detection of lipid peroxidation and oxidative DNA damage in non-alcoholic fatty liver diseases. J Hepatol. 2002;37(1):56–62.
Khetani SR, Szulgit G, Del Rio JA, et al. Exploring interactions between rat hepatocytes and nonparenchymal cells using gene expression profiling. Hepatology. 2004;40(3):545–54.
Zhang Z, Li XJ, Liu Y, et al. Recombinant human decorin inhibits cell proliferation and downregulates TGF-beta1 production in hypertrophic scar fibroblasts. Burns. 2007;33(5):634–41.
Kalamajski S, Aspberg A, Lindblom K, et al. Asporin competes with decorin for collagen binding, binds calcium and promotes osteoblast collagen mineralization. Biochem J. 2009;423(1):53–9.
Chen AA, Khetani SR, Lee S, et al. Modulation of hepatocyte phenotype in vitro via chemomechanical tuning of polyelectrolyte multilayers. Biomaterials. 2009;30(6):1113–20.
Wang Y, Lu C, Wei H, et al. Hepatopoietin interacts directly with COP9 signalosome and regulates AP-1 activity. FEBS Lett. 2004;572(1–3):85–91.
Pearce C, Hayden RE, Bunce CM, et al. Analysis of the role of COP9 signalosome (CSN) subunits in K562; the first link between CSN and autophagy. BMC Cell Biol. 2009;10:31.
Kato JY, Yoneda-Kato N. Mammalian COP9 signalosome. Genes Cells. 2009;14(11):1209–25.
Laplante JM, O'Rourke F, Lu X, et al. Cloning of human Ca2+ homoeostasis endoplasmic reticulum protein (CHERP): regulated expression of antisense cDNA depletes CHERP, inhibits intracellular Ca2+ mobilization and decreases cell proliferation. Biochem J. 2000;348(Pt 1):189–99.
Vallee RB, Williams JC, Varma D, et al. Dynein: an ancient motor protein involved in multiple modes of transport. J Neurobiol. 2004;58(2):189–200.
Gennerich A, Vale RD. Walking the walk: how kinesin and dynein coordinate their steps. Curr Opin Cell Biol. 2009;21(1):59–67.
Feldstein AE, Canbay A, Angulo P, et al. Hepatocyte apoptosis and fas expression are prominent features of human nonalcoholic steatohepatitis. Gastroenterology. 2003;125(2):437–43.
Malhi H, Bronk SF, Werneburg NW, et al. Free fatty acids induce JNK-dependent hepatocyte lipoapoptosis. J Biol Chem. 2006;281(17):12093–101.
Li Z, Oben JA, Yang S, et al. Norepinephrine regulates hepatic innate immune system in leptin-deficient mice with nonalcoholic steatohepatitis. Hepatology. 2004;40(2):434–41.
Li Z, Soloski MJ, Diehl AM. Dietary factors alter hepatic innate immune system in mice with nonalcoholic fatty liver disease. Hepatology. 2005;42(4):880–5.
den Boer MA, Voshol PJ, Schroder-van der Elst JP, et al. Endogenous interleukin-10 protects against hepatic steatosis but does not improve insulin sensitivity during high-fat feeding in mice. Endocrinology. 2006;147(10):4553–8.
Chan SS, Schedlich LJ, Twigg SM, et al. Inhibition of adipocyte differentiation by insulin-like growth factor-binding protein-3. Am J Physiol Endocrinol Metab. 2009;296(4):E654–63.
Silha JV, Gui Y, Murphy LJ. Impaired glucose homeostasis in insulin-like growth factor-binding protein-3-transgenic mice. Am J Physiol Endocrinol Metab. 2002;283(5):E937–45.
Chan SS, Twigg SM, Firth SM, et al. Insulin-like growth factor binding protein-3 leads to insulin resistance in adipocytes. J Clin Endocrinol Metab. 2005;90(12):6588–95.
Oh Y, Muller HL, Lamson G, et al. Insulin-like growth factor (IGF)-independent action of IGF-binding protein-3 in Hs578T human breast cancer cells. Cell surface binding and growth inhibition. J Biol Chem. 1993;268(20):14964–71.
Rajah R, Valentinis B, Cohen P. Insulin-like growth factor (IGF)-binding protein-3 induces apoptosis and mediates the effects of transforming growth factor-beta1 on programmed cell death through a p53- and IGF-independent mechanism. J Biol Chem. 1997;272(18):12181–8.
Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008;454(7203):455–62.
Hirsch C, Gauss R, Horn SC, et al. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 2009;458(7237):453–60.
Wei Y, Wang D, Pagliassotti MJ. Saturated fatty acid-mediated endoplasmic reticulum stress and apoptosis are augmented by trans-10, cis-12-conjugated linoleic acid in liver cells. Mol Cell Biochem. 2007;303(1–2):105–13.
Puri P, Mirshahi F, Cheung O, et al. Activation and dysregulation of the unfolded protein response in nonalcoholic fatty liver disease. Gastroenterology. 2008;134(2):568–76.
Ni M, Lee AS. ER chaperones in mammalian development and human diseases. FEBS Lett. 2007;581(19):3641–51.
Zhang L, Lai E, Teodoro T, et al. GRP78, but Not Protein-disulfide isomerase, partially reverses hyperglycemia-induced inhibition of insulin synthesis and secretion in pancreatic {beta}-cells. J Biol Chem. 2009;284(8):5289–98.
Kammoun HL, Chabanon H, Hainault I, et al. GRP78 expression inhibits insulin and ER stress-induced SREBP-1c activation and reduces hepatic steatosis in mice. J Clin Invest. 2009;119(5):1201–15.
Valero R, Marfany G, Gonzalez-Angulo O, et al. USP25, a novel gene encoding a deubiquitinating enzyme, is located in the gene-poor region 21q11.2. Genomics. 1999;62(3):395–405.
Valero R, Bayes M, Francisca Sanchez-Font M, et al. Characterization of alternatively spliced products and tissue-specific isoforms of USP28 and USP25. Genome Biol. 2001;2(10):RESEARCH0043.
Carlson MR, Zhang B, Fang Z, et al. Gene connectivity, function, and sequence conservation: predictions from modular yeast co-expression networks. BMC Genomics. 2006;7:40.
MacLennan NK, Dong J, Aten JE, et al. Weighted gene co-expression network analysis identifies biomarkers in glycerol kinase deficient mice. Mol Genet Metab. 2009;98(1–2):203–14.
Horvath S, Zhang B, Carlson M, et al. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc Natl Acad Sci U S A. 2006;103(46):17402–7.
Saris CG, Horvath S, van Vught PW, et al. Weighted gene co-expression network analysis of the peripheral blood from amyotrophic lateral sclerosis patients. BMC Genomics. 2009;10:405.
Sharma MR, Polavarapu R, Roseman D, et al. Transcriptional networks in a rat model for nonalcoholic fatty liver disease: a microarray analysis. Exp Mol Pathol. 2006;81(3):202–10.
Author information
Authors and Affiliations
Corresponding author
Additional information
This work was funded by a grant from the Biotechnology and Bioengineering Center of the Medical College of Wisconsin (SG and MO) and grant HL74168 from the Heart, Lung, and Blood Institute of the National Institutes of Health (MO).
Rights and permissions
About this article
Cite this article
Gawrieh, S., Baye, T.M., Carless, M. et al. Hepatic Gene Networks in Morbidly Obese Patients With Nonalcoholic Fatty Liver Disease. OBES SURG 20, 1698–1709 (2010). https://doi.org/10.1007/s11695-010-0171-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11695-010-0171-6