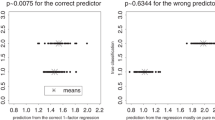
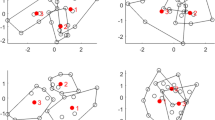
Abstract
Linear discriminant analysis (LDA) is a multivariate classification technique frequently applied to morphometric data in various biomedical disciplines. Canonical variate analysis (CVA), the generalization of LDA for multiple groups, is often used in the exploratory style of an ordination technique (a low-dimensional representation of the data). In the rare case when all groups have the same covariance matrix, maximum likelihood classification can be based on these linear functions. Both LDA and CVA require full-rank covariance matrices, which is usually not the case in modern morphometrics. When the number of variables is close to the number of individuals, groups appear separated in a CVA plot even if they are samples from the same population. Hence, reliable classification and assessment of group separation require many more organisms than variables. A simple alternative to CVA is the projection of the data onto the principal components of the group averages (between-group PCA). In contrast to CVA, these axes are orthogonal and can be computed even when the data are not of full rank, such as for Procrustes shape coordinates arising in samples of any size, and when covariance matrices are heterogeneous. In evolutionary quantitative genetics, the selection gradient is identical to the coefficient vector of a linear discriminant function between the populations before vs. after selection. When the measured variables are Procrustes shape coordinates, discriminant functions and selection gradients are vectors in shape space and can be visualized as shape deformations. Except for applications in quantitative genetics and in classification, however, discriminant functions typically offer no interpretation as biological factors.






Similar content being viewed by others
References
Adams, D. C., & Funk, D. J. (1997). Morphometric inferences on sibling species and sexual dimorphism in Neochlamisus bebbianae leaf beetles: Multivariate applications of the thin-plate spline. Systematic Biology, 46(1),180–194.
Adams, D. C., Rohlf, F. J., & Slice, D. E. (2004). Geometric morphometrics: Ten years of progress following the “revolution”. Italian Journal of Zoology, 71(9), 5–16.
Arnold, S. J., Bürger, R., Holenhole, P. A., Beverly, C. A., & Jones, A. G. (2008). Understanding the evolution and stability of the G-matrix. Evolution, 62, 2451–2461.
Arnold, S. J., Pfrender, M. E., & Jones, A. (2001). The adaptive landscape as a conceptual bridge between micro- and macroevolution. Genetica, 112–113, 9–32.
Barker, M., & Rayens, W. (2003). Partial least squares for discrimination. Journal of Chemometrics, 17, 166–173.
Blackith, R. E., & Reyment, R. A. (1971). Multivariate morphometrics. London: Academic Press.
Bookstein, F. (1991). Morphometric tools for landmark data: Geometry and biology. Cambridge, UK: Cambridge University Press.
Bookstein, F. (1996). Biometrics, biomathematics and the morphometric synthesis. Bulletin of Mathematical Biology, 58(2), 313–365.
Bookstein, F. L. (2002). Creases as morphometric characters. In N. MacLeod & P. Forey (Eds.), Morphology, shape, and phylogeny (pp. 139–174). London: Taylor and Francis.
Boulesteix, A.-L. (2005). A note on between-group PCA. International Journal of Pure and Applied Mathematics, 19, 359–366.
Bowman, C. E. (2009). Megavariate genetics: What you find is what you go looking for. Biological Theory, 4(1), 21–28.
Burnaby, T. P. (1966). Growth-invariant discrimination functions and generalized distances. Biometrics, 22, 96–110.
Campbell, N. A., & Atchley, W. R. (1981). The geometry of canonical variate analysis. Systematic Zoology, 30(3), 268–280.
Cardini, A., & O’Higgins, P. (2004). Patterns of morphological evolution in Marmota (Rodentia, Sciuridae): Geometric morphometrics of the cranium in the context of marmot phylogeny, ecology and conservation. Biological Journal of the Linnean Society, 82, 385–407.
Culhane, A. C., Perriere, G., Considine, E. C., Cotter, T. G., & Higgins, D. G. (2002). Between-group analysis of microarray data. Bioinformatics, 18(12), 1600–1608.
Dryden, I. L., & Mardia, K. V. (1998). Statistical shape analysis. New York: Wiley.
Duda, R. O., Hart, P. E., & Stork, D. G. (2000). Pattern classification. New York: Wiley-Interscience.
Dworkin, I., & Gibson, G. (2006). Epidermal growth factor receptor and transforming growth factor-beta signaling contributes to variation for wing shape in Drosophila melanogaster. Genetics, 173(3), 1417–1431.
Fisher, R. A. (1936). The use of multiple measurements in taxonomic problems. Annals of Eugenics, 7, 179–188.
Fisher, R. A. (1938). The statistical utilization of multiple measurements. Annals of Eugenics, 8, 376–386.
Flury, L., Boukai, B., & Flury, B. D. (1997). The discrimination subspace model. Journal of the American Statistical Association, 92(438), 758–766.
Friedman, J. H. (1989). Regularized discriminant analysis. Journal of the American Statistical Association, 84(405), 165–175.
Gunz, P., Mitteroecker, P., & Bookstein, F. L. (2005). Semilandmarks in three dimensions. In: D. E. Slice (Ed.), Modern morphometrics in p hysical anthropology (pp. 73–98). New York: Kluwer Academic/Plenum Publishers.
Hallgrimsson, B., Brown, J. J., Ford-Hutchinson, A. F., Sheets, H. D., Zelditch, M. L., & Jirik, F. R. (2006). The brachymorph mouse and the developmental-genetic basis for canalization and morphological integration. Evolution & Development, 8(1), 61–73.
Hallgrimsson, B., Jamniczky, H., Young, N. M., Rolian, C., Parson, T. E., Boughner, J. C., et al. (2009). Deciphering the palimpsest: Studying the relationship between morphological integration and phenotypic covariation. Evolutionary Biology, 36(4), 355–376.
Harvati, K. (2003). Quantitative analysis of Neanderthal temporal bone morphology using three-dimensional geometric morphometrics. American Journal of Physical Anthropology, 120(4), 323–338.
Henderson, H. V., & Searle, S. R. (1981). On deriving the inverse of a sum of matrices. SIAM Review, 23(1), 53–60.
Huberty, C. J., & Barton, R. M. (1989). An introduction to discriminant analysis. Measurement & Evaluation in Counseling & Development, 22, 158–168.
Huberty, C. J., & Curry, A. R. (1978). Linear versus quadratic multivariate classification. Multivariate Behavioral Research, 13(2), 237–245.
Huttegger, S., & Mitteroecker, P. (submitted). Invariance and meaningfulness in phenotype spaces. Evolutionary Biology.
Johnson, R. A., & Wichern, D. W. (1998). Applied multivariate statistical analysis. Upper Saddle River, NJ: Prentice-Hall.
Jolliffe, I. T. (2002). Principal component analysis. New York: Springer.
Kemsley, E. K. (1996). Discriminant analysis of high-dimensional data: A comparison of principal components analysis and partial least squares data reduction methods. Chemometrics and Intelligent Laboratory Systems, 33, 47–61.
Klingenberg, C. P., & Monteiro, L. R. (2005). Distances and directions in multidimensional shape spaces: Implications for morphometric applications. Systematic Biology, 54(4), 678–688.
Klingenberg, C. P., & Spence, J. R. (1993). Heterochrony and allometry: Lessons from the water strider genus Limnoporus. Evolution, 47, 1834–1853.
Lande, R. (1979). Quantitative genetic analysis of multivariate evolution, applied to brain: Body size allometry. Evolution, 33, 402–416.
Lande, R., & Arnold, S. J. (1983). The measurement of selection on correlated character. Evolution, 37, 1210–1226.
Lawing, A. M., & Polly, P. D. (2010). Geometric morphometrics: Recent applications to the study of evolution and development. Journal of Zoology, 280, 1–7.
Leinonen, T., Cano, J. M., Makinen, H., & Merila, J. (2006). Contrasting patterns of body shape and neutral genetic divergence in marine and lake populations of threespine sticklebacks. Journal of Evolutionary Biology, 19(6), 1803–1812.
Lele, S., & Richtsmeier, J. T. (1991). Euclidean distance matrix analysis: A coordinate free approach for comparing biological shapes using landmark data. American Journal of Physical Anthropology, 86, 415–428.
Lynch, M., & Walsh, B. (1998). Genetics and analysis of quantitative traits. Sunderland, MA: Sinauer Associates.
McLachlan, G.J. (2004). Discriminant analysis and statistical pattern recognition. New York: Wiley-Interscience.
MacLeod, N. (1999). Generalizing and extending the eigenshape method of shape visualization and analysis. Paleobiology, 25(1), 107–138.
MacLeod, N., O’Neill, M. A., & Walsh, S. A. (2005). A comparison between morphometric and artificial neural-net approaches to the automated species-recognition problem in systematics. In G. Curry & C. Humphries (Eds.), Biodiversity databases: From Cottage industry to industrial network. London: Taylor & Francis.
Mardia, K. V., Kent, J. T., & Bibby, J. M. (1979). Multivariate analysis. London: Academic Press.
Mitteroecker, P., & Bookstein, F. (2009). The ontogenetic trajectory of the phenotypic covariance matrix, with examples from craniofacial shape in rats and humans. Evolution, 63(3), 727–737.
Mitteroecker, P., & Bookstein, F. L. (2007). The conceptual and statistical relationship between modularity and morphological integration. Systematic Biology, 56(5), 818–836.
Mitteroecker, P., & Bookstein, F. L. (2008). The evolutionary role of modularity and integration in the hominoid cranium. Evolution, 62(4), 943–958.
Mitteroecker, P., & Gunz, P. (2009). Advances in geometric morphometrics. Evolutionary Biology, 36, 235–247.
Mitteroecker, P., Gunz, P., Bernhard, M., Schaefer, K., & Bookstein, F. (2004). Comparison of cranial ontogenetic trajectories among great apes and humans. Journal of Human Evolution, 46, 679–697.
Mitteroecker, P., Gunz, P., & Bookstein, F. L. (2005). Heterochrony and geometric morphometrics: A comparison of cranial growth in Pan paniscus versus Pan troglodytes. Evolution & Development, 7(3), 244–258.
Mitteroecker, P., & Huttegger, S. (2009). The concept of morphospaces in evolutionary and developmental biology: Mathematics and metaphors. Biological Theory, 4(1), 54–67.
Naylor, G. J. P., & Marcus, L. F. (1994). Identifying isolated shark teeth of the genus Carcharhinus to species: Relevance for tracking phyletic change through the fossil record. American Museum Novitates, 3109, 1–53.
O’Higgins, P. (2000). The study of morphological variation in the hominid fossil record: Biology, landmarks and geometry. Journal of Anatomy, 197, 103–120.
O’Neill, T. J. (1992). Error rates of non-Bayes classification rules and the robustness of Fisher’s linear discriminant function. Biometrica, 79(1), 177–184.
Parsons, T. E., Kristensen, E., Hornung, L., Diewert, V. M., Boyd, S. K., German, R. Z., et al. (2008). Phenotypic variability and craniofacial dysmorphology: Increased shape variance in a mouse model for cleft lip. Journal of Anatomy, 212(2), 135–143.
Pavlicev, M., Wagner, G., & Cheverud, J. M. (2009). Measuring evolutionary constraints through the dimensionality of the phenotype: Adjusted bootstrap method to estimate rank of phenotypic covariance matrices. Evolutionary Biology, 36, 339–353.
Rao, C. R. (1948). The utilization of multiple measurements in problems of biological classification. Journal of the Royal Statistical Society. Series B, 10(2), 159–203.
Rohlf, F. J., & Bookstein, F. (1987). A comment on shearing as a method for “size correction”. Systematic Zoology, 36, 356–367.
Rohlf, F. J, Loy, A., & Corti, M. (1996). Morphometric analysis of Old World Talpidae (Mammalia, Insectivora) using partial-warp scores. Systematic Biology, 45(3), 344–362.
Rohlf, F. J., & Marcus, L. F. (1993). A revolution in morphometrics. TREE, 8(4), 129–132.
Rohlf, F. J., & Slice, D. E. (1990). Extensions of the Procrustes method for the optimal superimposition of landmarks. Systematic Zoology, 39, 40–59.
Rosipal, R., & Krämer, N. (2006). Overview and recent advances in partial least squares. In C. Saunders, M. Grobelnik, S. Gunn, & J. Shawe-Taylor (Eds.), Subspace, latent structure and feature selection. Berlin: Springer.
Sheets, H. D., Covino, K. M., Panasiewicz, J. M., & Morris, S. R. (2006). Comparison of geometric morphometric outline methods in the discrimination of age-related differences in feather shape. Frontiers in Zoology, 3, 15.
Skinner, M. M., Gunz, P., Wood, B. A., Boesch, C., & Hublin, J. J. (2009). Discrimination of extant Pan species and subspecies using the enamel-dentine junction morphology of lower molars. American Journal of Physical Anthropology, 140(2), 234–243.
Slice, D. (2007). Geometric morphometrics. Annual Review of Anthropology, 36, 261–281.
Sneath, P., & Sokal, R. (1973). Numerical taxonomy: The principles and practice of numerical classification. San Francisco: W. H. Freeman.
Wright, S. (1932). General, group and special size factors. Genetics, 15, 603–619.
Zollikofer, C. P., & Ponce De Leon, M. S. (2002). Visualizing patterns of craniofacial shape variation in Homo sapiens. Proceedings of the Royal Society of London. Series B, Biological Sciences, 269(1493), 801–807.
Acknowledgments
We thank Philipp Gunz, Hans Nemeschkal, Dennis Slice, James Rohlf and two anonymous reviewers for helpful comments on the manuscript and for discussion.
Author information
Authors and Affiliations
Corresponding author
Appendix
Appendix
Figure 2 shows that the log likelihood surface for two groups A, B with the same covariance matrix \({\varvec{\Upsigma}}_A={\varvec{\Upsigma}}_B \equiv {\varvec{\Upsigma}}\) is a flat plane with a gradient vector \({\varvec{\Upsigma}}^{-1}({\varvec{\mu}}_A-{\varvec{\mu}}_B)\). To prove this, at least informally, consider \({\bf y}_i={\varvec{\Upsigma}}^{-1/2}{\bf x}_i\), so that the within-group covariance matrix of these transformed variables y is equal to the identity matrix. Then, still assuming equal covariance matrices, the log likelihood ratio, i.e., the exponent of (1), is half the difference between the squared Euclidean distances of x i to the two group means. This, in turn, is equal to the distance of x i from the midpoint of the group means \({{1}\over {2}}({\varvec{\mu}}_A+{\varvec{\mu}}_B)\), projected onto the vector of group mean differences \({\varvec{\mu}}_A-{\varvec{\mu}}_B\):
Apparently, likelihood ratios are then constant along lines orthogonal to the vector of group mean differences, and lines of constant log likelihood ratio are regularly spaced, constituting a flat log likelihood ratio surface with the gradient vector \({\varvec{\mu}}_A-{\varvec{\mu}}_B\). Since the multiplication by \({\varvec{\Upsigma}}^{1/2}\) is a linear transformation that preserves linear structures, the surface must be flat for the untransformed variables \({\bf x}_i={\varvec{\Upsigma}}^{1/2}{\bf y}_i\), too.
Let X be a mean-centered n × p data matrix for a sample consisting of two distinct groups with the group averages \(\bar{{\bf x}}_1, \bar{{\bf x}}_2\) and sample sizes \(n_1, n_2\) so that \(n=n_1+n_2\). The total sum of squares T = X′X can be decomposed into the pooled within-group sum of squares W and the between-group sum of squares B. The discriminant vector a usually is computed as the vector of group mean differences premultiplied by the inverse of W:
Consider the n-dimensional vector d with elements \(d_i=1/n_1\) if the ith specimen belongs to group 1 and \(d_i=-1/n_2\) if it belongs to group 2, so that \({\bf X}^{\prime}{\bf d}= \bar{\bf x}_1-\bar{\bf x}_2\) is the group mean difference. The multiple regression of this dummy variable on shape
gives a vector that is proportional to the linear discriminant vector. This implies that the discriminant function can also be computed by premultiplying the vector of group mean differences \({\bf g}=\bar{{\bf x}}_1-\bar{{\bf x}}_2\) by the inverse of the total sum of squares matrix T rather than the within-group sum of squares matrix W. To prove this, consider first that the between-group sum of squares matrix B has only one non-zero eigenvalue. The corresponding eigenvector is \({\bf e}={\bf g}/\|{\bf g}\|\) and the eigenvalue λ is the trace of B or \(\| \bf g\|^2/2\). Multiplying B with the vector of group mean differences thus gives Bg = λ g. Furthermore, the inverse of the sum of two matrices can be decomposed in the following way:
where A and C are invertible and B, C are arbitrary (rectangular) matrices (Henderson and Searle 1981). Let \({\bf A=W, B=g, C=}{\varvec{\lambda}}\), and D = g′, then
where ξ is a scalar. Another proof is given by Fisher (1938).
Discriminant functions are invariant to affine transformations of the variables, including separate changes of the scale of each variable. Let A be any regular p × p matrix; then XA is a linear transformation of the original variables X. The linear discriminant function for these transformed data is identical to the discriminant function of the original variables:
Several authors suggested visualizing the regression of shape on the discriminant scores Xa, that is
As the length of this vector is irrelevant, we can ignore the scalar quantity (a′X′Xa)−1 and replace (7) by the term
which is proportional to the vector of covariances between the shape variables and the discriminant score. When substituting a in (8) by (6) we get
where \(\zeta\) is a scalar value. In words, the vector of coefficients for the regression of X on the discriminant function is proportional to the mean difference vector.
Similarly, instead of a direct visualization of selection gradients, Klingenberg and Monteiro (2005) suggested visualizing the multivariate regression of shape on the scores along the selection gradient \({\varvec{\beta}}\). This vector of regression coefficients is proportional to the vector \(({\bf X}{\varvec{\beta}})^{\prime}{\bf X}\). Klingenberg and Monteiro noticed that this leads to a visualization of the scaled selection differential s. A proof follows from (9) when taking a as the selection gradient \({\varvec{\beta}}\) and \(\bar{{\bf x}}_1, \bar{{\bf x}}_2\) as the group means after and before selection.
Rights and permissions
About this article
Cite this article
Mitteroecker, P., Bookstein, F. Linear Discrimination, Ordination, and the Visualization of Selection Gradients in Modern Morphometrics. Evol Biol 38, 100–114 (2011). https://doi.org/10.1007/s11692-011-9109-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11692-011-9109-8